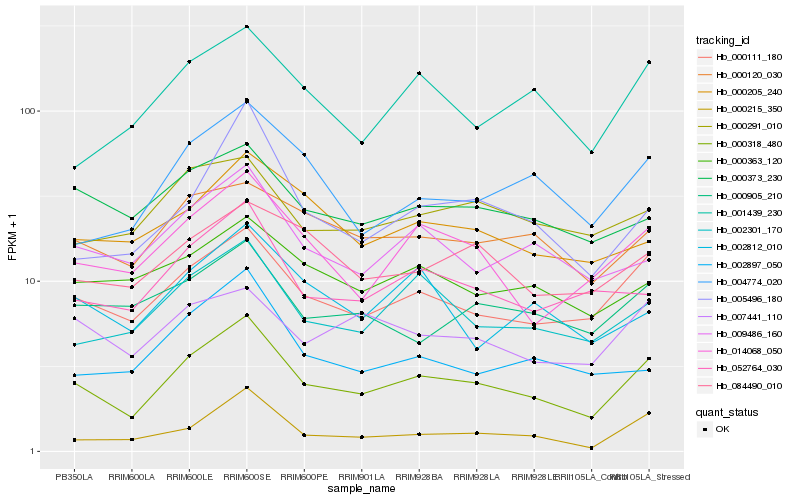
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000318_480 |
0.0 |
- |
- |
PREDICTED: RING-H2 finger protein ATL65 [Jatropha curcas] |
| 2 |
Hb_000111_180 |
0.0947739233 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 3 |
Hb_014068_050 |
0.1203686891 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 4 |
Hb_002301_170 |
0.1248313787 |
- |
- |
MAC/Perforin domain-containing protein [Theobroma cacao] |
| 5 |
Hb_052764_030 |
0.1289625803 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 6 |
Hb_009486_160 |
0.133001104 |
- |
- |
PREDICTED: metacaspase-1 [Jatropha curcas] |
| 7 |
Hb_000215_350 |
0.1330912096 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_005496_180 |
0.1361984737 |
- |
- |
PREDICTED: U-box domain-containing protein 4-like [Jatropha curcas] |
| 9 |
Hb_004774_020 |
0.1388087122 |
- |
- |
PREDICTED: PRA1 family protein B4-like [Camelina sativa] |
| 10 |
Hb_000120_030 |
0.1395264303 |
- |
- |
PREDICTED: uncharacterized protein LOC105630385 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001439_230 |
0.1413766434 |
- |
- |
PREDICTED: cystathionine gamma-synthase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000373_230 |
0.1431705108 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 13 |
Hb_007441_110 |
0.1434331077 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At3g58900-like [Jatropha curcas] |
| 14 |
Hb_084490_010 |
0.1444908649 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000905_210 |
0.145006699 |
- |
- |
PREDICTED: protein gar2 [Jatropha curcas] |
| 16 |
Hb_000291_010 |
0.146086935 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 17 |
Hb_002812_010 |
0.146196211 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000205_240 |
0.1462821856 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_002897_050 |
0.1467133361 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 20 |
Hb_000363_120 |
0.1472854575 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |