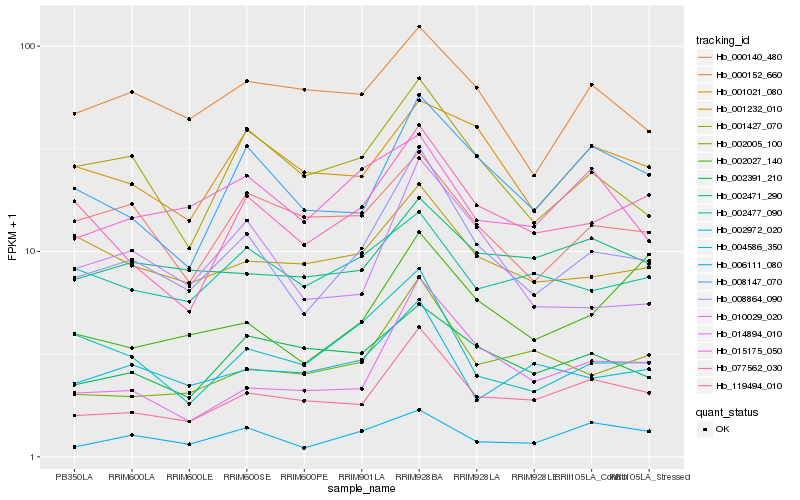
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008864_090 |
0.0 |
- |
- |
PREDICTED: plastid division protein PDV2-like [Jatropha curcas] |
| 2 |
Hb_119494_010 |
0.1136661576 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 3 |
Hb_001427_070 |
0.1210462342 |
- |
- |
PREDICTED: xaa-Pro dipeptidase [Jatropha curcas] |
| 4 |
Hb_010029_020 |
0.1229629274 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 [Jatropha curcas] |
| 5 |
Hb_077562_030 |
0.1278544599 |
- |
- |
PREDICTED: cytochrome P450 4X1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_008147_070 |
0.1286788459 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 7 |
Hb_014894_010 |
0.1326456444 |
- |
- |
hypothetical protein CICLE_v10001869mg [Citrus clementina] |
| 8 |
Hb_006111_080 |
0.1349752943 |
- |
- |
hypothetical protein JCGZ_18070 [Jatropha curcas] |
| 9 |
Hb_002972_020 |
0.135371112 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 10 |
Hb_002005_100 |
0.1356841024 |
- |
- |
- |
| 11 |
Hb_001232_010 |
0.1391469552 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 12 |
Hb_000140_480 |
0.1414300918 |
- |
- |
PREDICTED: uncharacterized protein LOC105637350 [Jatropha curcas] |
| 13 |
Hb_000152_660 |
0.1429865955 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 7 [Jatropha curcas] |
| 14 |
Hb_004586_350 |
0.1443425584 |
- |
- |
Phosphoribosylaminoimidazole-succinocarboxamide synthase [Morus notabilis] |
| 15 |
Hb_001021_080 |
0.1471034062 |
- |
- |
PREDICTED: autophagy-related protein 18a [Jatropha curcas] |
| 16 |
Hb_002391_210 |
0.1484748877 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |
| 17 |
Hb_002477_090 |
0.1498358044 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002471_290 |
0.15168352 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X1 [Jatropha curcas] |
| 19 |
Hb_015175_050 |
0.1517514373 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 10 isoform X3 [Jatropha curcas] |
| 20 |
Hb_002027_140 |
0.152441372 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.6 [Jatropha curcas] |