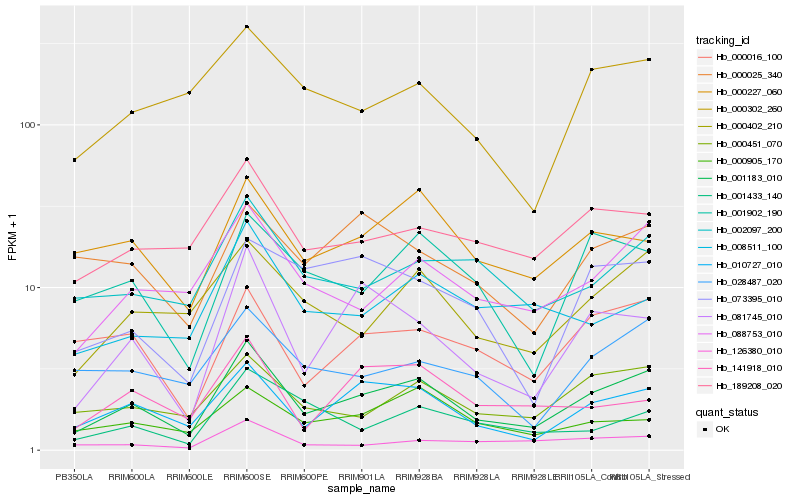
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001183_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_010727_010 |
0.1279627033 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 3 |
Hb_081745_010 |
0.1498353679 |
- |
- |
hypothetical protein CISIN_1g030953mg [Citrus sinensis] |
| 4 |
Hb_088753_010 |
0.1506579183 |
- |
- |
PREDICTED: copper amine oxidase 1-like [Vitis vinifera] |
| 5 |
Hb_000402_210 |
0.1556348838 |
- |
- |
hypothetical protein EUGRSUZ_B032971, partial [Eucalyptus grandis] |
| 6 |
Hb_000451_070 |
0.1557462584 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 7 |
Hb_000016_100 |
0.167097814 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP19 [Jatropha curcas] |
| 8 |
Hb_001902_190 |
0.168802013 |
- |
- |
PREDICTED: oxalate--CoA ligase [Jatropha curcas] |
| 9 |
Hb_000227_060 |
0.1718169104 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 10 |
Hb_189208_020 |
0.1721720649 |
- |
- |
LRX2, putative [Ricinus communis] |
| 11 |
Hb_002097_200 |
0.1739546888 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 12 |
Hb_126380_010 |
0.1775787828 |
desease resistance |
Gene Name: NB-ARC |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_028487_020 |
0.1789266589 |
- |
- |
hypothetical protein POPTR_0010s11880g [Populus trichocarpa] |
| 14 |
Hb_000905_170 |
0.1796219813 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 15 |
Hb_000302_260 |
0.1810091295 |
- |
- |
hypothetical protein [Bruguiera gymnorhiza] |
| 16 |
Hb_008511_100 |
0.1854351056 |
- |
- |
hypothetical protein POPTR_0009s11860g [Populus trichocarpa] |
| 17 |
Hb_073395_010 |
0.1870099007 |
- |
- |
- |
| 18 |
Hb_000025_340 |
0.1895653063 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62350 [Jatropha curcas] |
| 19 |
Hb_141918_010 |
0.19025962 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 20 |
Hb_001433_140 |
0.1937321459 |
- |
- |
PREDICTED: phospholipase D p1 [Jatropha curcas] |