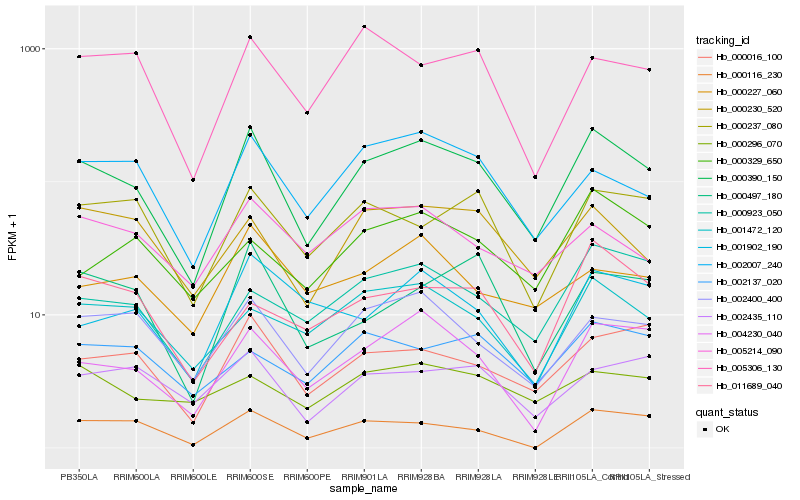
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000390_150 |
0.0 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105632469 [Jatropha curcas] |
| 2 |
Hb_004230_040 |
0.134352006 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002400_400 |
0.142947456 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X3 [Jatropha curcas] |
| 4 |
Hb_000016_100 |
0.1450701232 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP19 [Jatropha curcas] |
| 5 |
Hb_002007_240 |
0.1470370992 |
- |
- |
PREDICTED: F-box protein At1g55000 [Jatropha curcas] |
| 6 |
Hb_000230_520 |
0.1502074782 |
- |
- |
PREDICTED: serine incorporator 3 [Jatropha curcas] |
| 7 |
Hb_001472_120 |
0.1533088026 |
- |
- |
- |
| 8 |
Hb_000923_050 |
0.1561799485 |
- |
- |
PREDICTED: transcription factor bHLH113 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000237_080 |
0.158980432 |
transcription factor |
TF Family: PHD |
PREDICTED: chromatin structure-remodeling complex subunit RSC1 [Jatropha curcas] |
| 10 |
Hb_000497_180 |
0.1594190442 |
- |
- |
hypothetical protein POPTR_0001s12710g [Populus trichocarpa] |
| 11 |
Hb_002435_110 |
0.1646055091 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromodomain-helicase-DNA-binding protein 1-like [Jatropha curcas] |
| 12 |
Hb_000116_230 |
0.1647626819 |
- |
- |
- |
| 13 |
Hb_005214_090 |
0.1678049317 |
- |
- |
PREDICTED: uncharacterized protein LOC105636015 [Jatropha curcas] |
| 14 |
Hb_001902_190 |
0.1690872977 |
- |
- |
PREDICTED: oxalate--CoA ligase [Jatropha curcas] |
| 15 |
Hb_000227_060 |
0.1699578558 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 16 |
Hb_000329_650 |
0.1707881955 |
- |
- |
hypothetical protein POPTR_0014s056001g, partial [Populus trichocarpa] |
| 17 |
Hb_000296_070 |
0.171085212 |
- |
- |
hypothetical protein B456_003G048700 [Gossypium raimondii] |
| 18 |
Hb_005306_130 |
0.1711994973 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 19 |
Hb_011689_040 |
0.1714495065 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |
| 20 |
Hb_002137_020 |
0.1717641008 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |