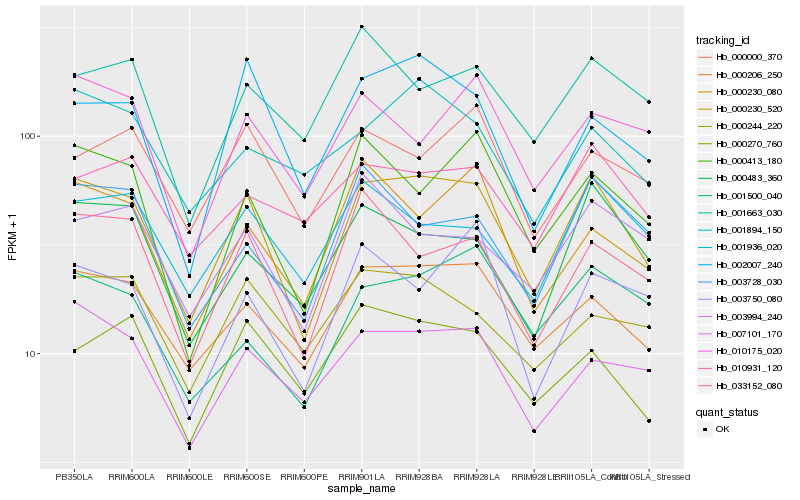
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000230_520 |
0.0 |
- |
- |
PREDICTED: serine incorporator 3 [Jatropha curcas] |
| 2 |
Hb_000206_250 |
0.0905442844 |
- |
- |
PREDICTED: UBP1-associated protein 2A-like [Jatropha curcas] |
| 3 |
Hb_033152_080 |
0.1078752982 |
- |
- |
RNA-binding protein Nova-1, putative [Ricinus communis] |
| 4 |
Hb_007101_170 |
0.1116038758 |
- |
- |
hypothetical protein POPTR_0010s10490g [Populus trichocarpa] |
| 5 |
Hb_000483_360 |
0.1120596109 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 6 |
Hb_000413_180 |
0.1123077427 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 7 |
Hb_002007_240 |
0.1174539736 |
- |
- |
PREDICTED: F-box protein At1g55000 [Jatropha curcas] |
| 8 |
Hb_001936_020 |
0.1178311958 |
- |
- |
PREDICTED: phosphatidylinositol 3-kinase, root isoform isoform X1 [Jatropha curcas] |
| 9 |
Hb_003728_030 |
0.1196990453 |
- |
- |
o-sialoglycoprotein endopeptidase, putative [Ricinus communis] |
| 10 |
Hb_000270_760 |
0.122124732 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_010931_120 |
0.1225787606 |
- |
- |
myosin heavy chain-related family protein [Populus trichocarpa] |
| 12 |
Hb_000230_080 |
0.1231474537 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 13 |
Hb_003994_240 |
0.1234022588 |
- |
- |
Mediator subunit 8 isoform 3 [Theobroma cacao] |
| 14 |
Hb_001894_150 |
0.1240855341 |
- |
- |
nucleoredoxin, putative [Ricinus communis] |
| 15 |
Hb_000244_220 |
0.1240881153 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_000000_370 |
0.1244172245 |
transcription factor |
TF Family: BES1 |
PREDICTED: protein BRASSINAZOLE-RESISTANT 1 [Jatropha curcas] |
| 17 |
Hb_001500_040 |
0.1245277319 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-1, mitochondrial isoform X2 [Jatropha curcas] |
| 18 |
Hb_010175_020 |
0.1269015346 |
- |
- |
PREDICTED: cysteine desulfurase, mitochondrial isoform X1 [Jatropha curcas] |
| 19 |
Hb_001663_030 |
0.1271781707 |
- |
- |
PREDICTED: peroxisomal membrane protein 13 [Jatropha curcas] |
| 20 |
Hb_003750_080 |
0.1272219046 |
- |
- |
PREDICTED: DNA repair protein complementing XP-C cells homolog [Jatropha curcas] |