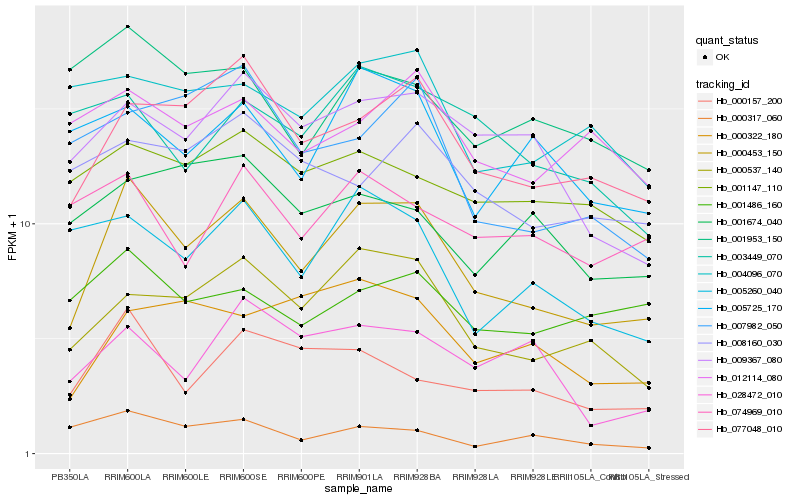
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000453_150 |
0.0 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 2 |
Hb_077048_010 |
0.1247676823 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000537_140 |
0.1292820912 |
- |
- |
lupus la ribonucleoprotein, putative [Ricinus communis] |
| 4 |
Hb_000157_200 |
0.1499871161 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 isoform X3 [Eucalyptus grandis] |
| 5 |
Hb_009367_080 |
0.152100987 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 42 [Jatropha curcas] |
| 6 |
Hb_000322_180 |
0.1625658122 |
- |
- |
recA family protein [Populus trichocarpa] |
| 7 |
Hb_005260_040 |
0.1641667336 |
- |
- |
PREDICTED: uncharacterized protein LOC105633768 isoform X1 [Jatropha curcas] |
| 8 |
Hb_005725_170 |
0.1680033692 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X2 [Jatropha curcas] |
| 9 |
Hb_001147_110 |
0.1693966889 |
- |
- |
PREDICTED: dynamin-2A-like [Jatropha curcas] |
| 10 |
Hb_028472_010 |
0.1724685685 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_007982_050 |
0.172581784 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2-like [Jatropha curcas] |
| 12 |
Hb_008160_030 |
0.1734962137 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_004096_070 |
0.1743103655 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_012114_080 |
0.1749809536 |
- |
- |
gamma-glutamylcysteine synthetase [Hevea brasiliensis] |
| 15 |
Hb_001953_150 |
0.1750322184 |
- |
- |
PREDICTED: protein DEK isoform X2 [Jatropha curcas] |
| 16 |
Hb_001486_160 |
0.1764865777 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 17 |
Hb_000317_060 |
0.1765166986 |
- |
- |
PREDICTED: uncharacterized protein LOC105647051 [Jatropha curcas] |
| 18 |
Hb_074969_010 |
0.1780451786 |
- |
- |
hypothetical protein JCGZ_23339 [Jatropha curcas] |
| 19 |
Hb_001674_040 |
0.1792024661 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 23 [Jatropha curcas] |
| 20 |
Hb_003449_070 |
0.1799507397 |
transcription factor |
TF Family: SNF2 |
DNA repair helicase rad5,16, putative [Ricinus communis] |