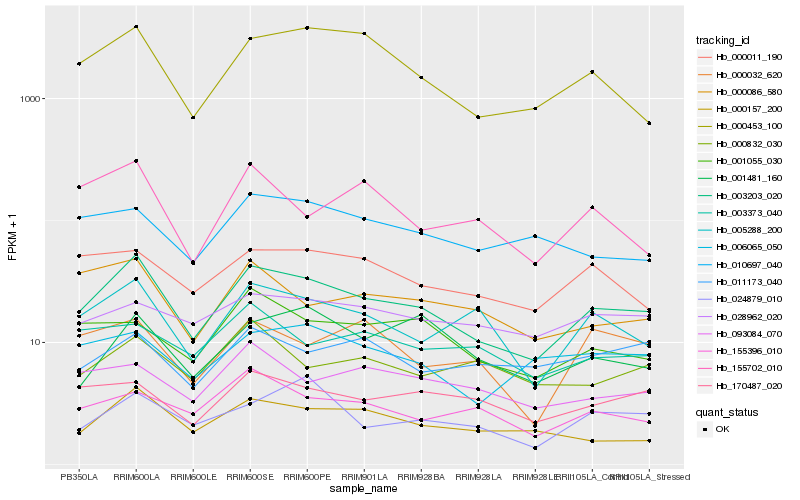
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003203_020 |
0.0 |
- |
- |
unknown [Lotus japonicus] |
| 2 |
Hb_005288_200 |
0.1334014776 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 3 |
Hb_000157_200 |
0.135668437 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 isoform X3 [Eucalyptus grandis] |
| 4 |
Hb_000453_100 |
0.1376131611 |
- |
- |
PREDICTED: metallothionein-like protein 1 [Sesamum indicum] |
| 5 |
Hb_000011_190 |
0.1490595356 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 6 [Jatropha curcas] |
| 6 |
Hb_000086_580 |
0.1494717438 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 7 |
Hb_155396_010 |
0.1507576527 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 8 |
Hb_006065_050 |
0.151987884 |
transcription factor |
TF Family: AP2 |
PREDICTED: floral homeotic protein APETALA 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_170487_020 |
0.1535895504 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 10 |
Hb_001055_030 |
0.1549476486 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 14 isoform X1 [Jatropha curcas] |
| 11 |
Hb_093084_070 |
0.1565165525 |
desease resistance |
Gene Name: Baculo_PEP_C |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 12 |
Hb_010697_040 |
0.1597044249 |
- |
- |
PREDICTED: protein GPR107 isoform X1 [Jatropha curcas] |
| 13 |
Hb_024879_010 |
0.1603723765 |
- |
- |
unknown [Glycine max] |
| 14 |
Hb_155702_010 |
0.1618627198 |
- |
- |
Tetraspanin-5 [Gossypium arboreum] |
| 15 |
Hb_000832_030 |
0.1619835035 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105634915 [Jatropha curcas] |
| 16 |
Hb_028962_020 |
0.1620706664 |
- |
- |
hypothetical protein POPTR_0016s00660g [Populus trichocarpa] |
| 17 |
Hb_000032_620 |
0.1621961839 |
- |
- |
hypothetical protein RCOM_1311830 [Ricinus communis] |
| 18 |
Hb_003373_040 |
0.162476217 |
- |
- |
hypothetical protein ARALYDRAFT_356474 [Arabidopsis lyrata subsp. lyrata] |
| 19 |
Hb_001481_160 |
0.1629137373 |
- |
- |
PREDICTED: RING-H2 finger protein ATL22-like [Populus euphratica] |
| 20 |
Hb_011173_040 |
0.1632116624 |
- |
- |
PREDICTED: protein ROOT PRIMORDIUM DEFECTIVE 1 [Populus euphratica] |