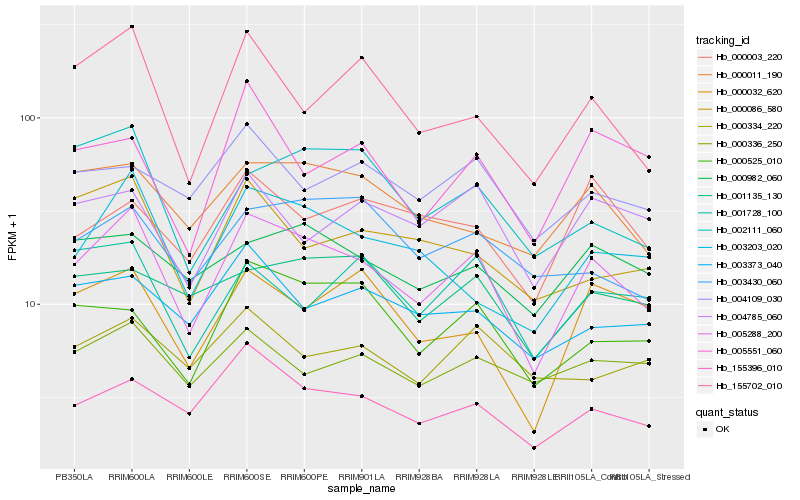
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005288_200 |
0.0 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 2 |
Hb_155396_010 |
0.1181064338 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 3 |
Hb_155702_010 |
0.1295094914 |
- |
- |
Tetraspanin-5 [Gossypium arboreum] |
| 4 |
Hb_003203_020 |
0.1334014776 |
- |
- |
unknown [Lotus japonicus] |
| 5 |
Hb_000525_010 |
0.137991258 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 6 |
Hb_000011_190 |
0.1383185892 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 6 [Jatropha curcas] |
| 7 |
Hb_000032_620 |
0.1408758995 |
- |
- |
hypothetical protein RCOM_1311830 [Ricinus communis] |
| 8 |
Hb_002111_060 |
0.1458759327 |
- |
- |
PREDICTED: toll-like receptor 13 [Jatropha curcas] |
| 9 |
Hb_000982_060 |
0.1460566087 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 10 |
Hb_003373_040 |
0.1478918151 |
- |
- |
hypothetical protein ARALYDRAFT_356474 [Arabidopsis lyrata subsp. lyrata] |
| 11 |
Hb_004785_060 |
0.1486034703 |
- |
- |
PREDICTED: SKP1-interacting partner 15 [Jatropha curcas] |
| 12 |
Hb_000086_580 |
0.150344454 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 13 |
Hb_003430_060 |
0.1506730285 |
- |
- |
PREDICTED: potassium transporter 23 isoform X2 [Elaeis guineensis] |
| 14 |
Hb_000336_250 |
0.1507936796 |
transcription factor |
TF Family: mTERF |
hypothetical protein B456_002G027700 [Gossypium raimondii] |
| 15 |
Hb_004109_030 |
0.151472938 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000003_220 |
0.1516815043 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 [Jatropha curcas] |
| 17 |
Hb_001728_100 |
0.1526427527 |
- |
- |
hypothetical protein JCGZ_03429 [Jatropha curcas] |
| 18 |
Hb_005551_060 |
0.1534497096 |
- |
- |
PREDICTED: uncharacterized protein LOC105635819 [Jatropha curcas] |
| 19 |
Hb_001135_130 |
0.1538610592 |
- |
- |
hypothetical protein CISIN_1g0171752mg, partial [Citrus sinensis] |
| 20 |
Hb_000334_220 |
0.1552039136 |
- |
- |
hypothetical protein JCGZ_16772 [Jatropha curcas] |