Hb_000453_100
Information
| Type | - |
|---|---|
| Description | - |
| Location | Contig453: 51772-52895 |
| Sequence |   |
Annotation
kegg
| ID | pop:POPTR_0019s13150g |
|---|---|
| description | hypothetical protein |
nr
| ID | XP_011089462.1 |
|---|---|
| description | PREDICTED: metallothionein-like protein 1 [Sesamum indicum] |
swissprot
| ID | Q0IMG5 |
|---|---|
| description | Metallothionein-like protein 4A OS=Oryza sativa subsp. japonica GN=MT4A PE=2 SV=1 |
trembl
| ID | Q7XHJ3 |
|---|---|
| description | Metallothionein-like protein (Fragment) OS=Quercus robur PE=2 SV=1 |
Gene Ontology
| ID | GO:0046872 |
|---|---|
| description | metallothionein-like protein |
Full-length cDNA clone information
| cDNA+EST (Sanger&Illumina) (ID:Location) |
PASA_asmbl_43257: 51785-52772 |
|---|---|
| cDNA (Sanger) (ID:Location) |
005_J18.ab1: 51850-52772 , 008_F09.ab1: 51851-52772 , 008_J12.ab1: 51992-52772 , 010_P03.ab1: 51848-52772 , 011_N02.ab1: 51848-52772 , 014_G09.ab1: 51850-52772 , 016_B24.ab1: 51986-52772 , 016_C17.ab1: 51848-52772 , 017_C03.ab1: 52482-52772 , 019_C19.ab1: 51848-52772 , 024_A19.ab1: 52506-52772 , 025_G02.ab1: 51944-52771 , 025_L10.ab1: 51848-52772 , 027_B11.ab1: 52503-52772 , 031_P11.ab1: 51905-52772 , 038_F12.ab1: 51785-52771 , 041_A14.ab1: 52130-52654 , 041_M07.ab1: 51944-52771 , 041_O18.ab1: 52494-52772 , 047_M04.ab1: 51944-52771 , 048_G13.ab1: 51992-52772 , 049_P20.ab1: 51932-52766 , 052_K19.ab1: 51842-52772 |
Similar expressed genes (Top20)
Gene co-expression network
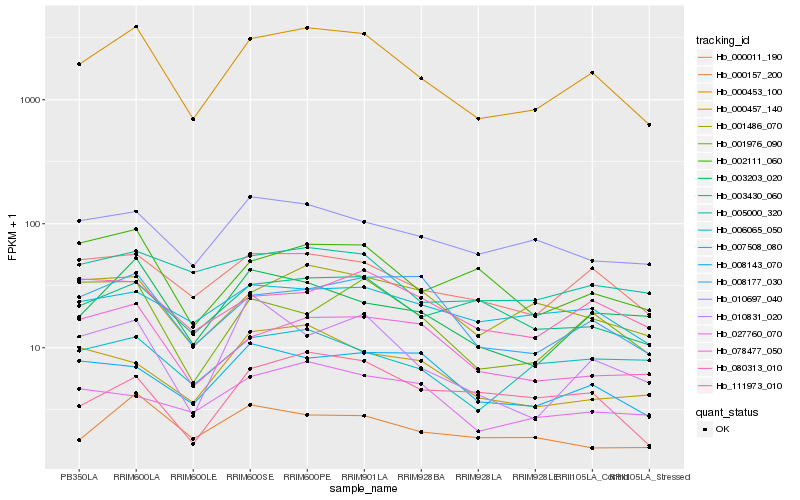
Expression pattern by RNA-Seq analysis
| RRIM600_Latex | RRIM600_Bark | RRIM600_Leaf | RRIM600_Petiole | PB350_Latex | RRIM901_Latex |
|---|---|---|---|---|---|
| 3887.93 | 3100.65 | 696.536 | 3805.31 | 1930.47 | 3406.77 |
| RRII105_Latex_C | RRII105_Latex_S | RRIM928_Latex | RRIM928_Bark | RRIM928_Leaf | |
| 1652.66 | 626.237 | 701.886 | 1485.01 | 828.077 |

