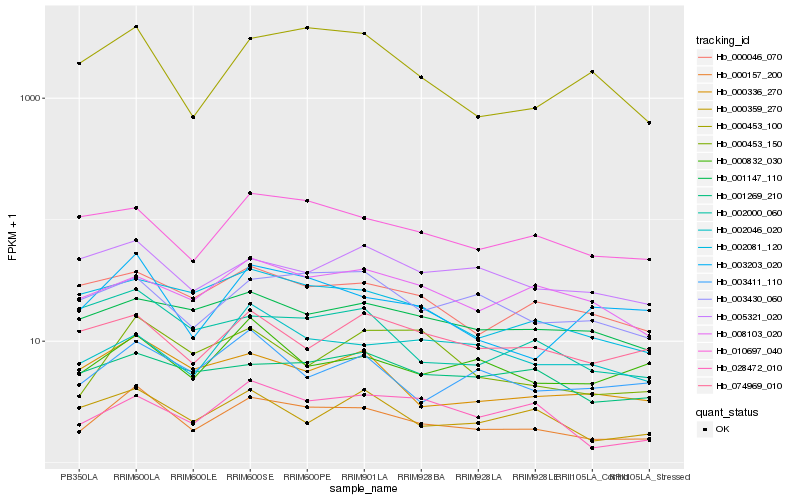
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000157_200 |
0.0 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 isoform X3 [Eucalyptus grandis] |
| 2 |
Hb_003203_020 |
0.135668437 |
- |
- |
unknown [Lotus japonicus] |
| 3 |
Hb_003430_060 |
0.1361724317 |
- |
- |
PREDICTED: potassium transporter 23 isoform X2 [Elaeis guineensis] |
| 4 |
Hb_000336_270 |
0.1365499958 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_010697_040 |
0.1376577555 |
- |
- |
PREDICTED: protein GPR107 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002081_120 |
0.1399634534 |
- |
- |
PREDICTED: magnesium transporter MRS2-3 [Jatropha curcas] |
| 7 |
Hb_000046_070 |
0.1434695246 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 8 |
Hb_000453_100 |
0.1481955379 |
- |
- |
PREDICTED: metallothionein-like protein 1 [Sesamum indicum] |
| 9 |
Hb_003411_110 |
0.148255164 |
- |
- |
PREDICTED: uncharacterized protein LOC105642013 [Jatropha curcas] |
| 10 |
Hb_008103_020 |
0.1483799023 |
- |
- |
hypothetical protein POPTR_0003s20310g [Populus trichocarpa] |
| 11 |
Hb_000832_030 |
0.1495248342 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105634915 [Jatropha curcas] |
| 12 |
Hb_000453_150 |
0.1499871161 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 13 |
Hb_028472_010 |
0.1500661716 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_001147_110 |
0.1501823521 |
- |
- |
PREDICTED: dynamin-2A-like [Jatropha curcas] |
| 15 |
Hb_000359_270 |
0.1511218992 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_001269_210 |
0.1512429785 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g76280 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005321_020 |
0.1518488844 |
- |
- |
splicing factor-related family protein [Populus trichocarpa] |
| 18 |
Hb_002000_060 |
0.1541200495 |
- |
- |
PREDICTED: protein LIKE COV 2 [Jatropha curcas] |
| 19 |
Hb_002046_020 |
0.1543829597 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 20 |
Hb_074969_010 |
0.1555702882 |
- |
- |
hypothetical protein JCGZ_23339 [Jatropha curcas] |