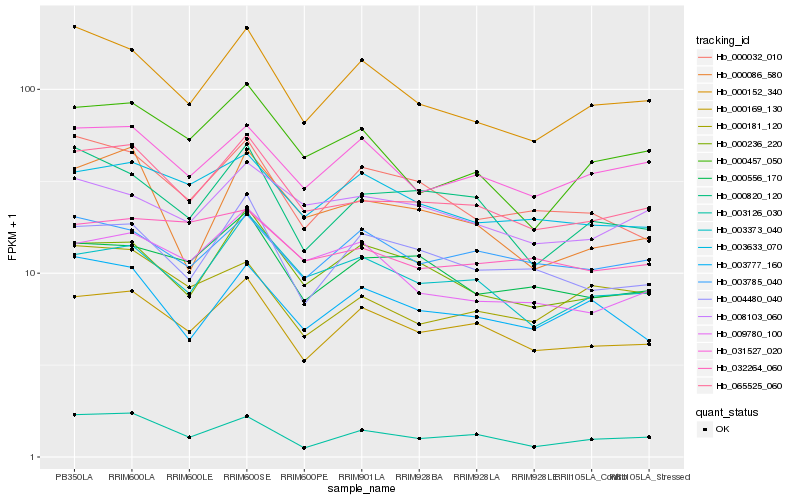
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_065525_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000169_130 |
0.069985993 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 3 |
Hb_003373_040 |
0.0809677717 |
- |
- |
hypothetical protein ARALYDRAFT_356474 [Arabidopsis lyrata subsp. lyrata] |
| 4 |
Hb_000152_340 |
0.086348497 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 5 |
Hb_000556_170 |
0.088105861 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 6 |
Hb_031527_020 |
0.0888133051 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B isoform X1 [Jatropha curcas] |
| 7 |
Hb_000236_220 |
0.0894407693 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 8 |
Hb_003633_070 |
0.0897094156 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 9 |
Hb_000820_120 |
0.0906467209 |
- |
- |
PREDICTED: uncharacterized protein LOC105639821 [Jatropha curcas] |
| 10 |
Hb_000457_050 |
0.0917849894 |
- |
- |
PREDICTED: tether containing UBX domain for GLUT4 [Jatropha curcas] |
| 11 |
Hb_000086_580 |
0.0922091404 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 12 |
Hb_008103_060 |
0.0927460766 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |
| 13 |
Hb_000032_010 |
0.0940422591 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 14 |
Hb_004480_040 |
0.0940789581 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor homolog 1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_032264_060 |
0.0971008486 |
- |
- |
hypothetical protein JCGZ_02506 [Jatropha curcas] |
| 16 |
Hb_003785_040 |
0.0972068866 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_003126_030 |
0.0989360163 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003777_160 |
0.0991268583 |
transcription factor |
TF Family: bZIP |
PREDICTED: G-box-binding factor 4 isoform X1 [Jatropha curcas] |
| 19 |
Hb_009780_100 |
0.1000529421 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 20 |
Hb_000181_120 |
0.1010385504 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |