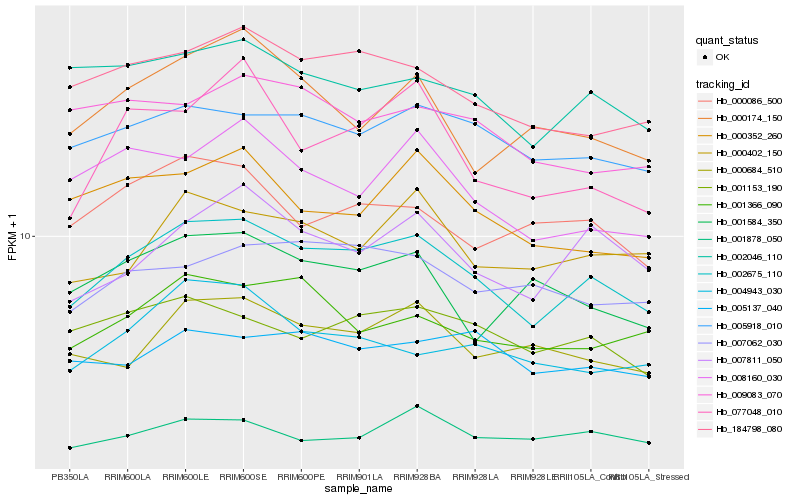
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002675_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634948 [Jatropha curcas] |
| 2 |
Hb_184798_080 |
0.0818764361 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 3 |
Hb_005918_010 |
0.0879229964 |
- |
- |
PREDICTED: protein ROOT HAIR DEFECTIVE 3-like [Jatropha curcas] |
| 4 |
Hb_007811_050 |
0.0891250486 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase NAK [Jatropha curcas] |
| 5 |
Hb_002046_110 |
0.0923254914 |
- |
- |
PREDICTED: TBC1 domain family member 15 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001153_190 |
0.094448202 |
- |
- |
PREDICTED: DNA-directed RNA polymerase IV subunit 1 [Jatropha curcas] |
| 7 |
Hb_000684_510 |
0.094886763 |
- |
- |
run and tbc1 domain containing 3, plant, putative [Ricinus communis] |
| 8 |
Hb_000402_150 |
0.0950845226 |
- |
- |
PREDICTED: uncharacterized protein LOC105644650 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001366_090 |
0.0957224651 |
transcription factor |
TF Family: NAC |
hypothetical protein RCOM_1341380 [Ricinus communis] |
| 10 |
Hb_007062_030 |
0.0991614918 |
- |
- |
hypothetical protein VITISV_039033 [Vitis vinifera] |
| 11 |
Hb_000086_500 |
0.0995439719 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_008160_030 |
0.0997274349 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_000352_260 |
0.1011452334 |
- |
- |
PREDICTED: WD repeat-containing protein LWD1 [Jatropha curcas] |
| 14 |
Hb_004943_030 |
0.1029161518 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |
| 15 |
Hb_000174_150 |
0.1039336452 |
- |
- |
stearoyl-acyl-carrier protein desaturase [Ricinus communis] |
| 16 |
Hb_005137_040 |
0.1042582962 |
- |
- |
hypothetical protein JCGZ_12324 [Jatropha curcas] |
| 17 |
Hb_077048_010 |
0.1050431436 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001878_050 |
0.1051968442 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 19 |
Hb_009083_070 |
0.1053802255 |
- |
- |
PREDICTED: uncharacterized protein LOC105639870 [Jatropha curcas] |
| 20 |
Hb_001584_350 |
0.1055307287 |
- |
- |
hypothetical protein JCGZ_23178 [Jatropha curcas] |