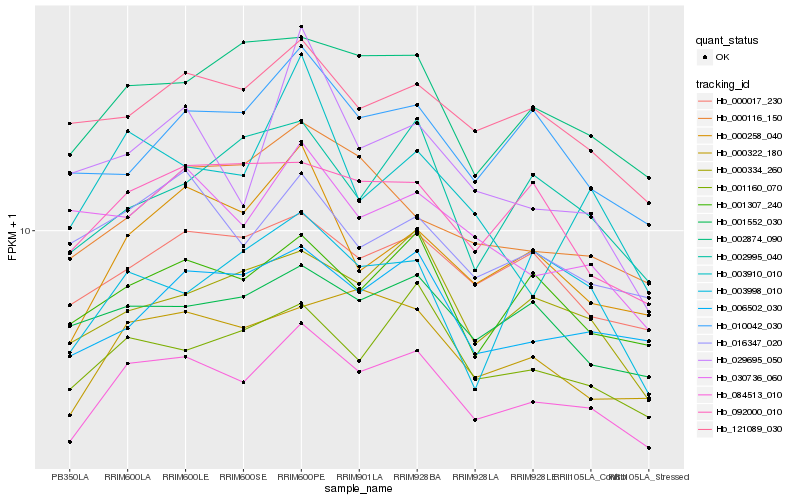
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_084513_010 |
0.0 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 5 [Jatropha curcas] |
| 2 |
Hb_000258_040 |
0.1243157099 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 3 |
Hb_000322_180 |
0.1255532377 |
- |
- |
recA family protein [Populus trichocarpa] |
| 4 |
Hb_001160_070 |
0.1319525553 |
- |
- |
PREDICTED: uncharacterized protein LOC105632163 [Jatropha curcas] |
| 5 |
Hb_002874_090 |
0.1358843127 |
- |
- |
PREDICTED: uncharacterized protein LOC105637452 [Jatropha curcas] |
| 6 |
Hb_001307_240 |
0.1377456485 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000334_260 |
0.1393610538 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 8 |
Hb_006502_030 |
0.1413341633 |
- |
- |
PREDICTED: probable kinetochore protein ndc80 [Jatropha curcas] |
| 9 |
Hb_000017_230 |
0.1469798817 |
- |
- |
hypothetical protein JCGZ_20558 [Jatropha curcas] |
| 10 |
Hb_092000_010 |
0.1495668305 |
- |
- |
kelch repeat-containing F-box family protein [Populus trichocarpa] |
| 11 |
Hb_121089_030 |
0.1523599135 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 12 |
Hb_003998_010 |
0.1527111322 |
- |
- |
PREDICTED: protein BRASSINOSTEROID INSENSITIVE 1 [Jatropha curcas] |
| 13 |
Hb_002995_040 |
0.1531249592 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase dyrk1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000116_150 |
0.1534439815 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 15 |
Hb_010042_030 |
0.1540223774 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine diphosphorylase 1 [Jatropha curcas] |
| 16 |
Hb_029695_050 |
0.1563644713 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 1 [Jatropha curcas] |
| 17 |
Hb_016347_020 |
0.1563754305 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like isoform X1 [Populus euphratica] |
| 18 |
Hb_001552_030 |
0.1565817323 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_003910_010 |
0.1576022742 |
- |
- |
PREDICTED: L-ascorbate oxidase homolog [Jatropha curcas] |
| 20 |
Hb_030736_060 |
0.1595178745 |
- |
- |
PREDICTED: lysM domain-containing GPI-anchored protein 1 [Jatropha curcas] |