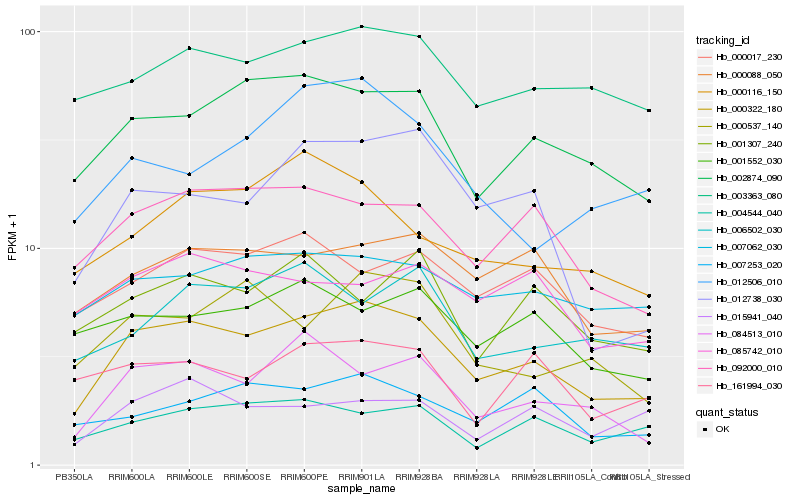
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000322_180 |
0.0 |
- |
- |
recA family protein [Populus trichocarpa] |
| 2 |
Hb_002874_090 |
0.1087813698 |
- |
- |
PREDICTED: uncharacterized protein LOC105637452 [Jatropha curcas] |
| 3 |
Hb_092000_010 |
0.115211346 |
- |
- |
kelch repeat-containing F-box family protein [Populus trichocarpa] |
| 4 |
Hb_000088_050 |
0.1234725201 |
- |
- |
PREDICTED: threonylcarbamoyladenosine tRNA methylthiotransferase [Jatropha curcas] |
| 5 |
Hb_084513_010 |
0.1255532377 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 5 [Jatropha curcas] |
| 6 |
Hb_003363_080 |
0.1286992511 |
- |
- |
eukaryotic translation elongation factor 1B gamma-subunit [Hevea brasiliensis] |
| 7 |
Hb_000017_230 |
0.1314618683 |
- |
- |
hypothetical protein JCGZ_20558 [Jatropha curcas] |
| 8 |
Hb_012738_030 |
0.1341268715 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007062_030 |
0.1341953212 |
- |
- |
hypothetical protein VITISV_039033 [Vitis vinifera] |
| 10 |
Hb_007253_020 |
0.134773378 |
- |
- |
PREDICTED: putative deoxyribonuclease TATDN1 [Jatropha curcas] |
| 11 |
Hb_004544_040 |
0.1363563626 |
- |
- |
hypothetical protein PHAVU_010G103000g [Phaseolus vulgaris] |
| 12 |
Hb_000537_140 |
0.136848463 |
- |
- |
lupus la ribonucleoprotein, putative [Ricinus communis] |
| 13 |
Hb_000116_150 |
0.1375240876 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 14 |
Hb_001552_030 |
0.1408283901 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_006502_030 |
0.1418456751 |
- |
- |
PREDICTED: probable kinetochore protein ndc80 [Jatropha curcas] |
| 16 |
Hb_015941_040 |
0.1427417106 |
- |
- |
L-galactono-1,4-lactone dehydrogenase, mitochondrial [Cucumis sativus] |
| 17 |
Hb_161994_030 |
0.1462810846 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_001307_240 |
0.14692324 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_012506_010 |
0.1479057523 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Jatropha curcas] |
| 20 |
Hb_085742_010 |
0.1480697827 |
- |
- |
hypothetical protein POPTR_0005s05170g [Populus trichocarpa] |