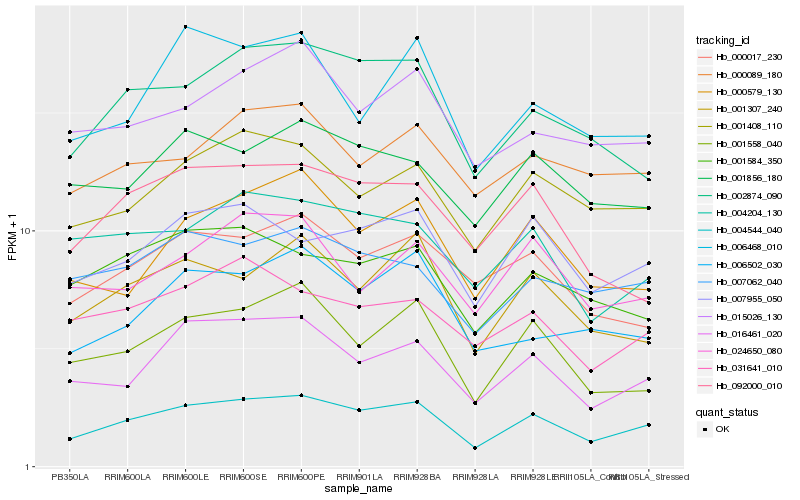
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004544_040 |
0.0 |
- |
- |
hypothetical protein PHAVU_010G103000g [Phaseolus vulgaris] |
| 2 |
Hb_016461_020 |
0.0827565568 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007955_050 |
0.0878590729 |
- |
- |
PREDICTED: multiple RNA-binding domain-containing protein 1 [Jatropha curcas] |
| 4 |
Hb_001408_110 |
0.0884396129 |
- |
- |
hypothetical protein EUGRSUZ_I02762 [Eucalyptus grandis] |
| 5 |
Hb_002874_090 |
0.0919208767 |
- |
- |
PREDICTED: uncharacterized protein LOC105637452 [Jatropha curcas] |
| 6 |
Hb_001558_040 |
0.0926902281 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 2 [Jatropha curcas] |
| 7 |
Hb_001307_240 |
0.0955599655 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000579_130 |
0.1001415399 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_007062_040 |
0.1010888536 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 10 |
Hb_004204_130 |
0.1026356454 |
- |
- |
PREDICTED: uncharacterized protein LOC103930912 [Pyrus x bretschneideri] |
| 11 |
Hb_001856_180 |
0.1034745742 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 12 |
Hb_031641_010 |
0.1044696103 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_015026_130 |
0.1047135244 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 14 |
Hb_000017_230 |
0.1057269703 |
- |
- |
hypothetical protein JCGZ_20558 [Jatropha curcas] |
| 15 |
Hb_001584_350 |
0.105971973 |
- |
- |
hypothetical protein JCGZ_23178 [Jatropha curcas] |
| 16 |
Hb_000089_180 |
0.1066401914 |
- |
- |
PREDICTED: homoserine kinase-like [Jatropha curcas] |
| 17 |
Hb_006502_030 |
0.1069303233 |
- |
- |
PREDICTED: probable kinetochore protein ndc80 [Jatropha curcas] |
| 18 |
Hb_092000_010 |
0.1072987494 |
- |
- |
kelch repeat-containing F-box family protein [Populus trichocarpa] |
| 19 |
Hb_024650_080 |
0.1074808669 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g09030 [Jatropha curcas] |
| 20 |
Hb_006468_010 |
0.1075076997 |
- |
- |
PREDICTED: eukaryotic translation initiation factor isoform 4G-1 isoform X1 [Jatropha curcas] |