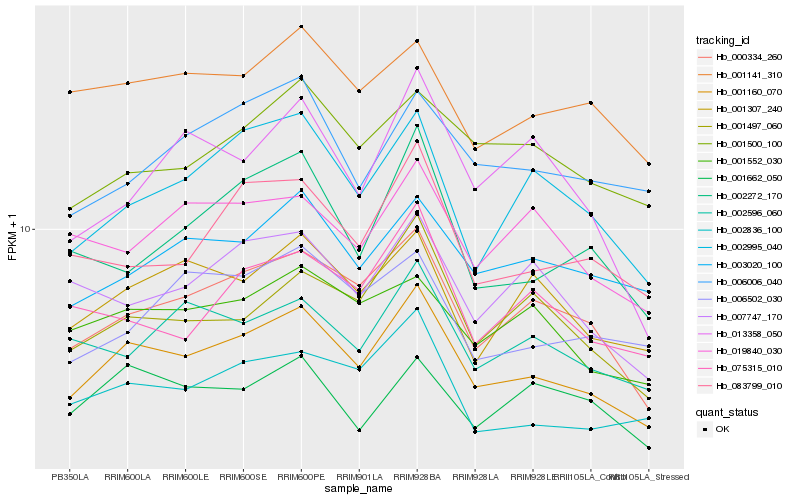
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001160_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632163 [Jatropha curcas] |
| 2 |
Hb_000334_260 |
0.0793923478 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 3 |
Hb_002995_040 |
0.0970370297 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase dyrk1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001497_060 |
0.1002800633 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 5 |
Hb_083799_010 |
0.10400741 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 6 |
Hb_001307_240 |
0.1042236241 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002836_100 |
0.104560942 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002272_170 |
0.1077274471 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase-like protein 1 [Jatropha curcas] |
| 9 |
Hb_001552_030 |
0.1082646793 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_007747_170 |
0.1101410391 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 11 |
Hb_003020_100 |
0.1108736068 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 12 |
Hb_002596_060 |
0.1116536896 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001141_310 |
0.115090981 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 14 |
Hb_075315_010 |
0.1158631151 |
- |
- |
ornithine aminotransferase [Camellia sinensis] |
| 15 |
Hb_001500_100 |
0.1171672263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_006006_040 |
0.1179623784 |
- |
- |
S-methyl-5-thioribose kinase isoform 2 [Theobroma cacao] |
| 17 |
Hb_006502_030 |
0.1182164854 |
- |
- |
PREDICTED: probable kinetochore protein ndc80 [Jatropha curcas] |
| 18 |
Hb_019840_030 |
0.1193389786 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 19 |
Hb_013358_050 |
0.1198163339 |
- |
- |
NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_001662_050 |
0.1208562053 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |