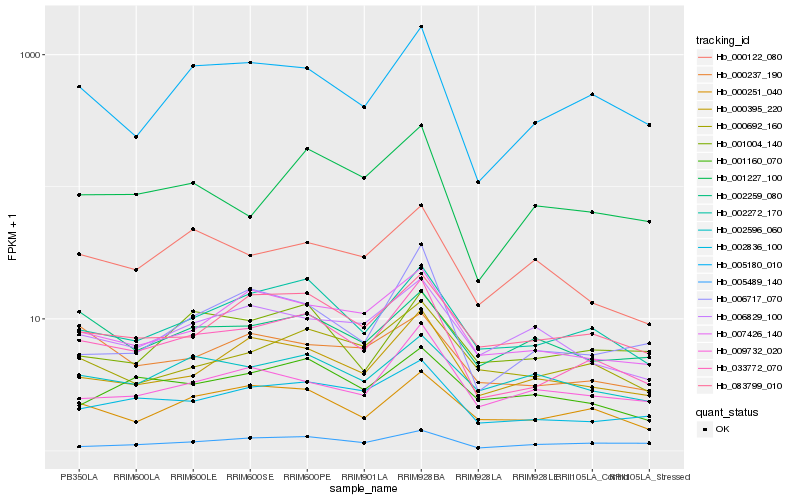
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033772_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633681 [Jatropha curcas] |
| 2 |
Hb_005180_010 |
0.1036163088 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 3 |
Hb_002272_170 |
0.1045055697 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase-like protein 1 [Jatropha curcas] |
| 4 |
Hb_002836_100 |
0.1134875019 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000395_220 |
0.1185364645 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 6 |
Hb_000251_040 |
0.1250031852 |
- |
- |
PREDICTED: DNA polymerase alpha catalytic subunit isoform X1 [Jatropha curcas] |
| 7 |
Hb_007426_140 |
0.1251974114 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 8 |
Hb_001227_100 |
0.1308068551 |
- |
- |
RecName: Full=Enolase 1; AltName: Full=2-phospho-D-glycerate hydro-lyase 1; AltName: Full=2-phosphoglycerate dehydratase 1; AltName: Allergen=Hev b 9 [Hevea brasiliensis] |
| 9 |
Hb_083799_010 |
0.1314511833 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 10 |
Hb_000692_160 |
0.1328256668 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 11 |
Hb_006717_070 |
0.1375789699 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor CPC-like [Jatropha curcas] |
| 12 |
Hb_009732_020 |
0.1400260951 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 13 |
Hb_005489_140 |
0.1431935623 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 14 |
Hb_000237_190 |
0.147952688 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000122_080 |
0.1481867814 |
- |
- |
PREDICTED: staphylococcal-like nuclease CAN2 [Jatropha curcas] |
| 16 |
Hb_002259_080 |
0.1505630647 |
- |
- |
protein transport protein sec23, putative [Ricinus communis] |
| 17 |
Hb_002596_060 |
0.1514276667 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001160_070 |
0.1516311035 |
- |
- |
PREDICTED: uncharacterized protein LOC105632163 [Jatropha curcas] |
| 19 |
Hb_001004_140 |
0.1547915408 |
- |
- |
hypothetical protein JCGZ_26893 [Jatropha curcas] |
| 20 |
Hb_006829_100 |
0.1550564902 |
- |
- |
conserved hypothetical protein [Ricinus communis] |