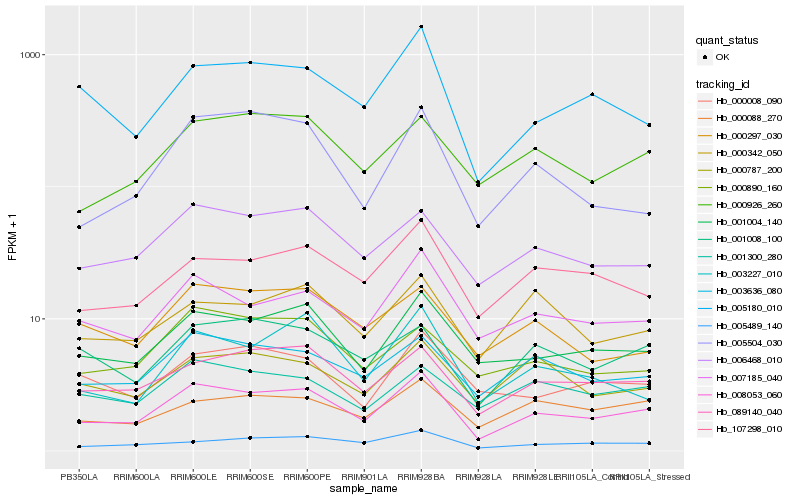
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008053_060 |
0.0 |
- |
- |
PREDICTED: protein PAIR1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001004_140 |
0.0991813055 |
- |
- |
hypothetical protein JCGZ_26893 [Jatropha curcas] |
| 3 |
Hb_089140_040 |
0.1023243355 |
- |
- |
PREDICTED: uncharacterized protein LOC105637966 [Jatropha curcas] |
| 4 |
Hb_007185_040 |
0.1079430203 |
- |
- |
PREDICTED: pyrroline-5-carboxylate reductase [Jatropha curcas] |
| 5 |
Hb_006468_010 |
0.1099189375 |
- |
- |
PREDICTED: eukaryotic translation initiation factor isoform 4G-1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003636_080 |
0.1121052063 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 [Jatropha curcas] |
| 7 |
Hb_005489_140 |
0.1165211267 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 8 |
Hb_107298_010 |
0.1178576691 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 9 |
Hb_005180_010 |
0.1199919265 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 10 |
Hb_000926_260 |
0.1204129885 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000088_270 |
0.1268498948 |
- |
- |
PREDICTED: uncharacterized protein LOC105637003 [Jatropha curcas] |
| 12 |
Hb_001008_100 |
0.1276860291 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 13 |
Hb_000890_160 |
0.1305559165 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 7 [Jatropha curcas] |
| 14 |
Hb_003227_010 |
0.1316396214 |
- |
- |
PREDICTED: dynamin-related protein 1E-like [Jatropha curcas] |
| 15 |
Hb_000297_030 |
0.132431999 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000787_200 |
0.133827649 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000008_090 |
0.1339343469 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 18 |
Hb_005504_030 |
0.1341777177 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2-like [Pyrus x bretschneideri] |
| 19 |
Hb_001300_280 |
0.1342307705 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein ucpB [Jatropha curcas] |
| 20 |
Hb_000342_050 |
0.1342358942 |
- |
- |
hypothetical protein POPTR_0006s24710g [Populus trichocarpa] |