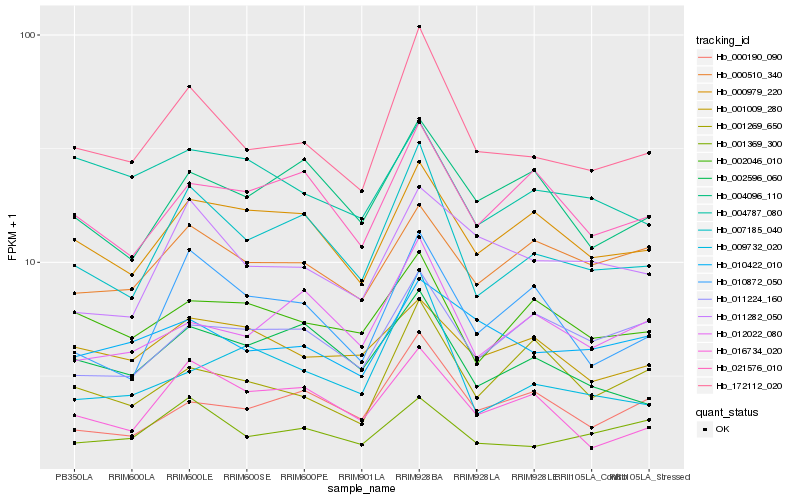
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_172112_020 |
0.0 |
- |
- |
PREDICTED: fumarylacetoacetase [Jatropha curcas] |
| 2 |
Hb_007185_040 |
0.0849649509 |
- |
- |
PREDICTED: pyrroline-5-carboxylate reductase [Jatropha curcas] |
| 3 |
Hb_010422_010 |
0.1112366249 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 4 |
Hb_000979_220 |
0.1143609012 |
- |
- |
PREDICTED: uncharacterized protein LOC105634085 isoform X1 [Jatropha curcas] |
| 5 |
Hb_011282_050 |
0.1188493832 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000190_090 |
0.1188592761 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 7 |
Hb_001369_300 |
0.1214028146 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 2-1-like [Jatropha curcas] |
| 8 |
Hb_016734_020 |
0.1229467336 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 9 |
Hb_012022_080 |
0.1231139816 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 10 |
Hb_021576_010 |
0.1240162067 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 11 |
Hb_002596_060 |
0.1257046823 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002046_010 |
0.1263434059 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_010872_050 |
0.1267176666 |
- |
- |
hypothetical protein B456_007G078100 [Gossypium raimondii] |
| 14 |
Hb_004096_110 |
0.1276684933 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 15 |
Hb_001269_650 |
0.1281401927 |
- |
- |
transporter, putative [Ricinus communis] |
| 16 |
Hb_009732_020 |
0.1287979319 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 17 |
Hb_000510_340 |
0.1304078236 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 18 |
Hb_001009_280 |
0.1328882077 |
- |
- |
PREDICTED: telomere length regulation protein TEL2 homolog [Jatropha curcas] |
| 19 |
Hb_004787_080 |
0.1366229375 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1B [Jatropha curcas] |
| 20 |
Hb_011224_160 |
0.136970417 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |