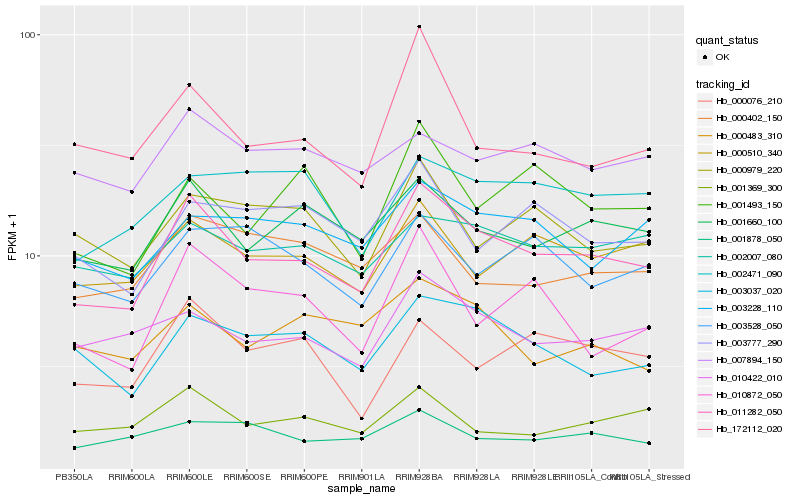
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011282_050 |
0.0 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_001660_100 |
0.0956412033 |
- |
- |
PREDICTED: glyoxylate reductase/hydroxypyruvate reductase [Jatropha curcas] |
| 3 |
Hb_010422_010 |
0.1060881693 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 4 |
Hb_003037_020 |
0.1066220351 |
- |
- |
hypothetical protein JCGZ_22910 [Jatropha curcas] |
| 5 |
Hb_000510_340 |
0.1069790915 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 6 |
Hb_002007_080 |
0.1138818762 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 7 |
Hb_001369_300 |
0.1181040836 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 2-1-like [Jatropha curcas] |
| 8 |
Hb_003228_110 |
0.1184960196 |
- |
- |
PREDICTED: uncharacterized protein LOC105637245 [Jatropha curcas] |
| 9 |
Hb_172112_020 |
0.1188493832 |
- |
- |
PREDICTED: fumarylacetoacetase [Jatropha curcas] |
| 10 |
Hb_000076_210 |
0.1196611704 |
- |
- |
PREDICTED: electron transfer flavoprotein subunit alpha, mitochondrial-like [Jatropha curcas] |
| 11 |
Hb_010872_050 |
0.1202970871 |
- |
- |
hypothetical protein B456_007G078100 [Gossypium raimondii] |
| 12 |
Hb_003777_290 |
0.1207011296 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 13 |
Hb_000979_220 |
0.1212297747 |
- |
- |
PREDICTED: uncharacterized protein LOC105634085 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001493_150 |
0.1221648484 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 15 |
Hb_002471_090 |
0.1223158705 |
- |
- |
PREDICTED: uncharacterized protein LOC105634169 [Jatropha curcas] |
| 16 |
Hb_001878_050 |
0.1226866482 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 17 |
Hb_000402_150 |
0.1230125395 |
- |
- |
PREDICTED: uncharacterized protein LOC105644650 isoform X2 [Jatropha curcas] |
| 18 |
Hb_007894_150 |
0.1234537851 |
- |
- |
hypothetical protein EUGRSUZ_F03462 [Eucalyptus grandis] |
| 19 |
Hb_003528_050 |
0.1242986476 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 20 |
Hb_000483_310 |
0.124838443 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4-like [Jatropha curcas] |