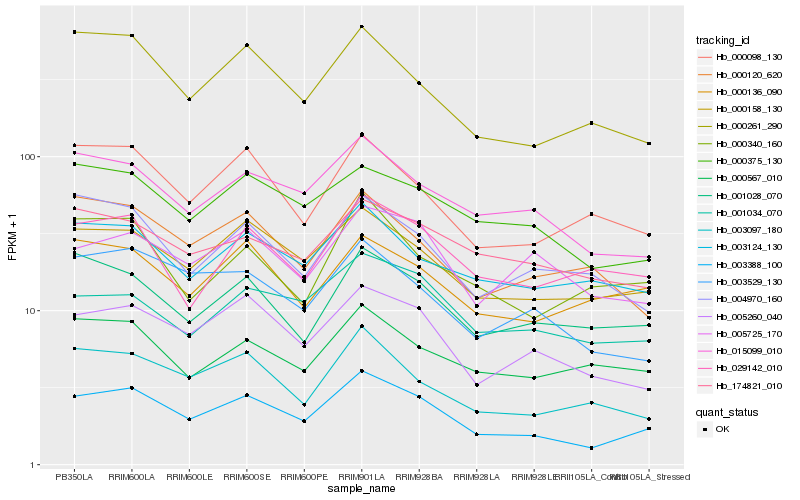
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003388_100 |
0.0 |
- |
- |
16S rRNA processing protein RimM [Psychrobacter phenylpyruvicus] |
| 2 |
Hb_000158_130 |
0.0861449749 |
- |
- |
PREDICTED: uncharacterized protein LOC105647950 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005260_040 |
0.0978195591 |
- |
- |
PREDICTED: uncharacterized protein LOC105633768 isoform X1 [Jatropha curcas] |
| 4 |
Hb_015099_010 |
0.103881431 |
- |
- |
PREDICTED: probable glucan 1,3-alpha-glucosidase [Jatropha curcas] |
| 5 |
Hb_000261_290 |
0.110840389 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 6 |
Hb_000375_130 |
0.1116649867 |
- |
- |
Leucine-rich repeat transmembrane protein kinase family protein isoform 1 [Theobroma cacao] |
| 7 |
Hb_003097_180 |
0.112249935 |
- |
- |
PREDICTED: crossover junction endonuclease MUS81 [Jatropha curcas] |
| 8 |
Hb_003529_130 |
0.1139749832 |
- |
- |
PREDICTED: ribosome biogenesis protein BOP1 homolog isoform X1 [Jatropha curcas] |
| 9 |
Hb_003124_130 |
0.1149440459 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000098_130 |
0.1152503786 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2A [Jatropha curcas] |
| 11 |
Hb_001028_070 |
0.1159358206 |
- |
- |
PREDICTED: WD repeat-containing protein 75 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001034_070 |
0.1164189043 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_029142_010 |
0.1205161771 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 14 |
Hb_000136_090 |
0.1213353342 |
- |
- |
programmed cell death protein, putative [Ricinus communis] |
| 15 |
Hb_000340_160 |
0.1249669412 |
- |
- |
PREDICTED: elongation factor Tu, mitochondrial [Jatropha curcas] |
| 16 |
Hb_005725_170 |
0.1252744921 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X2 [Jatropha curcas] |
| 17 |
Hb_174821_010 |
0.1261044195 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 14 [Jatropha curcas] |
| 18 |
Hb_000567_010 |
0.1268853568 |
- |
- |
PREDICTED: uncharacterized protein LOC105631110 [Jatropha curcas] |
| 19 |
Hb_000120_620 |
0.1280354178 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1a isoform X1 [Jatropha curcas] |
| 20 |
Hb_004970_160 |
0.1281052412 |
- |
- |
PREDICTED: THO complex subunit 5B-like isoform X2 [Jatropha curcas] |