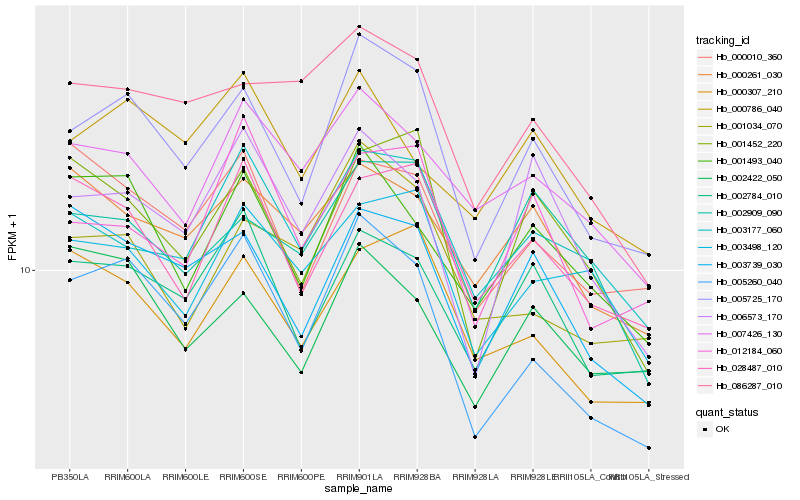
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005725_170 |
0.0 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X2 [Jatropha curcas] |
| 2 |
Hb_005260_040 |
0.0756831329 |
- |
- |
PREDICTED: uncharacterized protein LOC105633768 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002784_010 |
0.0848479377 |
- |
- |
DNA-directed RNA polymerase I subunit, putative [Ricinus communis] |
| 4 |
Hb_003739_030 |
0.0890022021 |
- |
- |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 [Jatropha curcas] |
| 5 |
Hb_001493_040 |
0.0925172844 |
- |
- |
PREDICTED: FACT complex subunit SPT16-like [Jatropha curcas] |
| 6 |
Hb_001452_220 |
0.0926313412 |
- |
- |
PREDICTED: molybdopterin biosynthesis protein CNX1 [Jatropha curcas] |
| 7 |
Hb_028487_010 |
0.0937293959 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000307_210 |
0.0937928582 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 9 |
Hb_001034_070 |
0.0975434536 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_003177_060 |
0.0983028393 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_006573_170 |
0.098940796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_012184_060 |
0.1001416324 |
- |
- |
PREDICTED: nuclear pore complex protein NUP1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002909_090 |
0.1006470782 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase DPE2 [Jatropha curcas] |
| 14 |
Hb_000010_360 |
0.1014966667 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 15 |
Hb_007426_130 |
0.1034045084 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 16 |
Hb_086287_010 |
0.1038830188 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003498_120 |
0.1042232224 |
- |
- |
hypothetical protein POPTR_0015s12430g, partial [Populus trichocarpa] |
| 18 |
Hb_002422_050 |
0.1044706816 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 5 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000261_030 |
0.1061366252 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 20 |
Hb_000786_040 |
0.1067432017 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 17 [Jatropha curcas] |