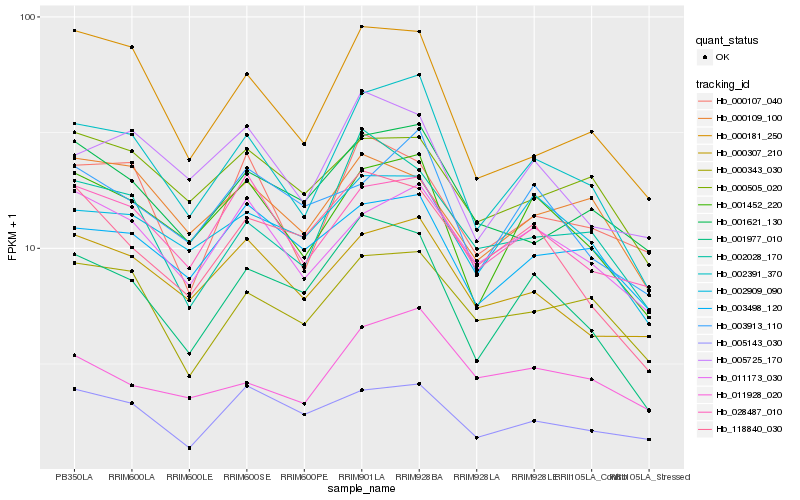
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002391_370 |
0.0 |
- |
- |
PREDICTED: aconitate hydratase 1 [Jatropha curcas] |
| 2 |
Hb_001452_220 |
0.0773546096 |
- |
- |
PREDICTED: molybdopterin biosynthesis protein CNX1 [Jatropha curcas] |
| 3 |
Hb_001977_010 |
0.0915111487 |
- |
- |
PREDICTED: uncharacterized protein LOC105636485 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000181_250 |
0.1010101154 |
- |
- |
PREDICTED: probable bifunctional methylthioribulose-1-phosphate dehydratase/enolase-phosphatase E1 [Jatropha curcas] |
| 5 |
Hb_002909_090 |
0.1075347462 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase DPE2 [Jatropha curcas] |
| 6 |
Hb_000307_210 |
0.1077113467 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 7 |
Hb_002028_170 |
0.1085548911 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 5 [Jatropha curcas] |
| 8 |
Hb_011173_030 |
0.1090453327 |
- |
- |
pumilio, putative [Ricinus communis] |
| 9 |
Hb_028487_010 |
0.1093364369 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 10 |
Hb_005725_170 |
0.1096646798 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X2 [Jatropha curcas] |
| 11 |
Hb_000343_030 |
0.1106747317 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 12 |
Hb_003913_110 |
0.1140681747 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 13 |
Hb_001621_130 |
0.1152359177 |
- |
- |
PREDICTED: pumilio homolog 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_003498_120 |
0.116269251 |
- |
- |
hypothetical protein POPTR_0015s12430g, partial [Populus trichocarpa] |
| 15 |
Hb_000107_040 |
0.118788225 |
- |
- |
nuclear pore complex protein nup153, putative [Ricinus communis] |
| 16 |
Hb_000505_020 |
0.1209559985 |
- |
- |
PREDICTED: protein S-acyltransferase 24 [Jatropha curcas] |
| 17 |
Hb_011928_020 |
0.1215724062 |
- |
- |
something about silencing protein sas10, putative [Ricinus communis] |
| 18 |
Hb_005143_030 |
0.1231904201 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 19 |
Hb_118840_030 |
0.1238655291 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 18 homolog [Jatropha curcas] |
| 20 |
Hb_000109_100 |
0.1238741998 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 [Jatropha curcas] |