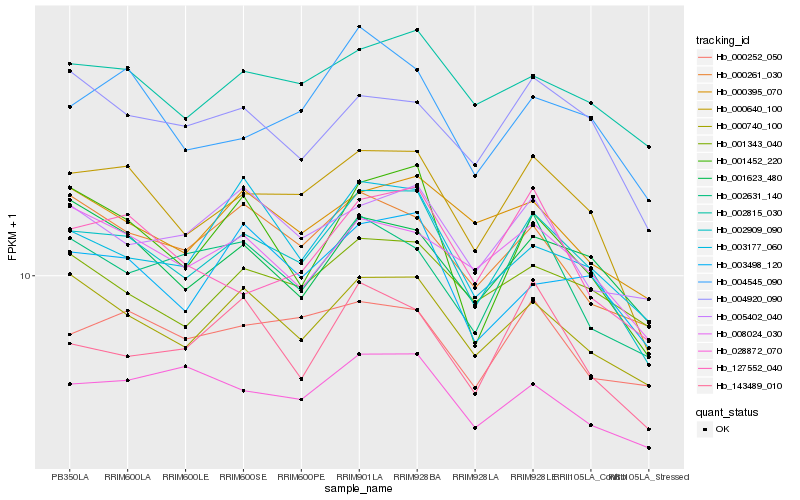
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002909_090 |
0.0 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase DPE2 [Jatropha curcas] |
| 2 |
Hb_000640_100 |
0.0494317141 |
- |
- |
PREDICTED: probable glutamyl endopeptidase, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_000740_100 |
0.0724059753 |
- |
- |
calpain, putative [Ricinus communis] |
| 4 |
Hb_002815_030 |
0.0820545184 |
- |
- |
hypothetical protein CISIN_1g0095162mg, partial [Citrus sinensis] |
| 5 |
Hb_001452_220 |
0.0826047546 |
- |
- |
PREDICTED: molybdopterin biosynthesis protein CNX1 [Jatropha curcas] |
| 6 |
Hb_003498_120 |
0.0851477207 |
- |
- |
hypothetical protein POPTR_0015s12430g, partial [Populus trichocarpa] |
| 7 |
Hb_004545_090 |
0.0877983597 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001343_040 |
0.0893738049 |
- |
- |
PREDICTED: uncharacterized protein LOC105638555 [Jatropha curcas] |
| 9 |
Hb_008024_030 |
0.0902771965 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105648348 [Jatropha curcas] |
| 10 |
Hb_000261_030 |
0.0905982428 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 11 |
Hb_143489_010 |
0.0913475361 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 50 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000395_070 |
0.0913615717 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_127552_040 |
0.0916894389 |
- |
- |
Uncharacterized protein L484_000643 [Morus notabilis] |
| 14 |
Hb_028872_070 |
0.0936423332 |
desease resistance |
Gene Name: DEAD |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase PRP1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001623_480 |
0.0937830731 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 13 [Jatropha curcas] |
| 16 |
Hb_005402_040 |
0.094223163 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 17 |
Hb_004920_090 |
0.0943492804 |
- |
- |
PREDICTED: probable glutamyl endopeptidase, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_003177_060 |
0.0943497982 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002631_140 |
0.0943584286 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 12, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000252_050 |
0.0944770147 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |