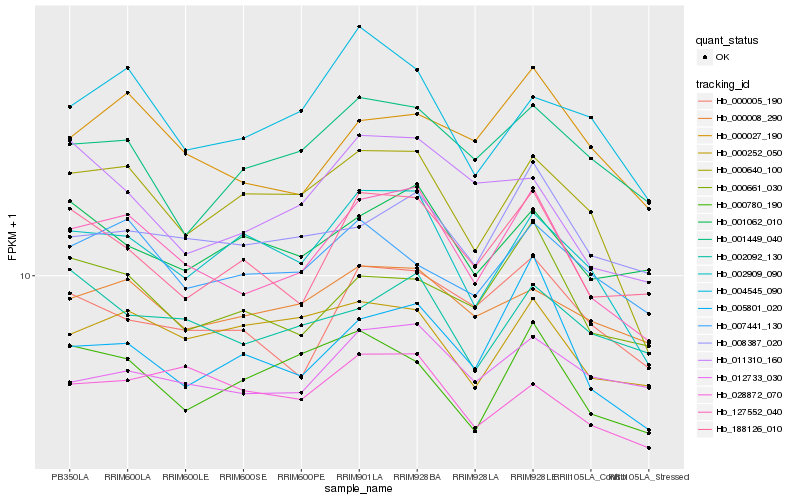
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_127552_040 |
0.0 |
- |
- |
Uncharacterized protein L484_000643 [Morus notabilis] |
| 2 |
Hb_188126_010 |
0.0916108729 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25-like isoform X3 [Jatropha curcas] |
| 3 |
Hb_002909_090 |
0.0916894389 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase DPE2 [Jatropha curcas] |
| 4 |
Hb_000640_100 |
0.0929990265 |
- |
- |
PREDICTED: probable glutamyl endopeptidase, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_005801_020 |
0.0969339025 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 6 |
Hb_000027_190 |
0.0980285496 |
- |
- |
PREDICTED: uncharacterized protein LOC105642057 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000661_030 |
0.1006571937 |
- |
- |
PREDICTED: uncharacterized protein LOC105650374 [Jatropha curcas] |
| 8 |
Hb_000005_190 |
0.1008975444 |
- |
- |
hypothetical protein JCGZ_05438 [Jatropha curcas] |
| 9 |
Hb_000008_290 |
0.1039202912 |
- |
- |
PREDICTED: protein SAND [Jatropha curcas] |
| 10 |
Hb_008387_020 |
0.105380205 |
- |
- |
PREDICTED: FHA domain-containing protein DDL isoform X1 [Jatropha curcas] |
| 11 |
Hb_012733_030 |
0.1069404278 |
- |
- |
hypothetical protein OsI_15243 [Oryza sativa Indica Group] |
| 12 |
Hb_000252_050 |
0.1076696602 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 13 |
Hb_000780_190 |
0.1081661297 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 2 [Jatropha curcas] |
| 14 |
Hb_004545_090 |
0.1084626786 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001449_040 |
0.1088071156 |
- |
- |
PREDICTED: SWR1 complex subunit 2 isoform X2 [Jatropha curcas] |
| 16 |
Hb_028872_070 |
0.1092197128 |
desease resistance |
Gene Name: DEAD |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase PRP1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002092_130 |
0.1098181649 |
- |
- |
PREDICTED: protein CASP-like [Oryza brachyantha] |
| 18 |
Hb_007441_130 |
0.1102243664 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_001062_010 |
0.1108144146 |
- |
- |
PREDICTED: CWF19-like protein 2 [Jatropha curcas] |
| 20 |
Hb_011310_160 |
0.1108771572 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |