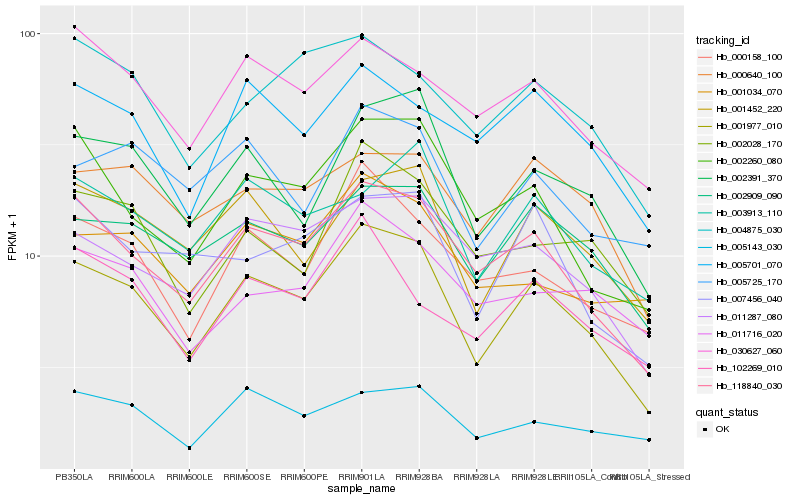
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001977_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636485 isoform X2 [Jatropha curcas] |
| 2 |
Hb_118840_030 |
0.0762705686 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 18 homolog [Jatropha curcas] |
| 3 |
Hb_002391_370 |
0.0915111487 |
- |
- |
PREDICTED: aconitate hydratase 1 [Jatropha curcas] |
| 4 |
Hb_001452_220 |
0.0999296421 |
- |
- |
PREDICTED: molybdopterin biosynthesis protein CNX1 [Jatropha curcas] |
| 5 |
Hb_002909_090 |
0.1044870789 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase DPE2 [Jatropha curcas] |
| 6 |
Hb_002260_080 |
0.1060134776 |
- |
- |
Aldehyde dehydrogenase family 6 member B2 [Morus notabilis] |
| 7 |
Hb_000158_100 |
0.1086141298 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_011287_080 |
0.112605492 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002028_170 |
0.1177557545 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 5 [Jatropha curcas] |
| 10 |
Hb_000640_100 |
0.1185724806 |
- |
- |
PREDICTED: probable glutamyl endopeptidase, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_005725_170 |
0.1198181755 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X2 [Jatropha curcas] |
| 12 |
Hb_007456_040 |
0.1204341968 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B [Jatropha curcas] |
| 13 |
Hb_005701_070 |
0.120746618 |
- |
- |
PREDICTED: uncharacterized TPR repeat-containing protein At1g05150-like [Jatropha curcas] |
| 14 |
Hb_003913_110 |
0.1211940357 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 15 |
Hb_030627_060 |
0.1212652262 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 16 |
Hb_005143_030 |
0.1226245645 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 17 |
Hb_001034_070 |
0.1235001496 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_102269_010 |
0.1243008376 |
- |
- |
hypothetical protein POPTR_0006s28940g [Populus trichocarpa] |
| 19 |
Hb_011716_020 |
0.1252308578 |
- |
- |
PREDICTED: uncharacterized protein LOC105637783 [Jatropha curcas] |
| 20 |
Hb_004875_030 |
0.1252939788 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Jatropha curcas] |