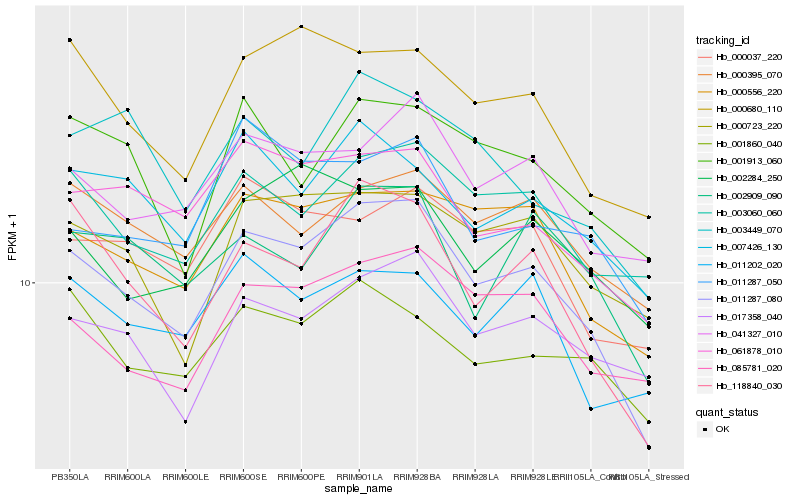
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011287_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_118840_030 |
0.0804024912 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 18 homolog [Jatropha curcas] |
| 3 |
Hb_000556_220 |
0.0847441618 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 2-like [Jatropha curcas] |
| 4 |
Hb_061878_010 |
0.094619827 |
- |
- |
hypothetical protein CISIN_1g0094072mg, partial [Citrus sinensis] |
| 5 |
Hb_000680_110 |
0.0960323152 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 6 |
Hb_041327_010 |
0.0965355055 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 7 |
Hb_003060_060 |
0.0966935499 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 8 |
Hb_085781_020 |
0.0974393954 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1 [Jatropha curcas] |
| 9 |
Hb_007426_130 |
0.1006069029 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 10 |
Hb_000723_220 |
0.1018685077 |
- |
- |
PREDICTED: uncharacterized protein LOC105642234 [Jatropha curcas] |
| 11 |
Hb_000037_220 |
0.1027459393 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 12 |
Hb_017358_040 |
0.1031806108 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |
| 13 |
Hb_000395_070 |
0.1032640586 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002909_090 |
0.103936669 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase DPE2 [Jatropha curcas] |
| 15 |
Hb_001860_040 |
0.105217371 |
- |
- |
PREDICTED: cleavage stimulation factor subunit 77 isoform X4 [Jatropha curcas] |
| 16 |
Hb_011287_050 |
0.1059501752 |
- |
- |
PREDICTED: uncharacterized protein LOC105644786 [Jatropha curcas] |
| 17 |
Hb_001913_060 |
0.1071381305 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 18 |
Hb_002284_250 |
0.1072349039 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 19 |
Hb_003449_070 |
0.10753889 |
transcription factor |
TF Family: SNF2 |
DNA repair helicase rad5,16, putative [Ricinus communis] |
| 20 |
Hb_011202_020 |
0.1081291223 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3 isoform X1 [Jatropha curcas] |