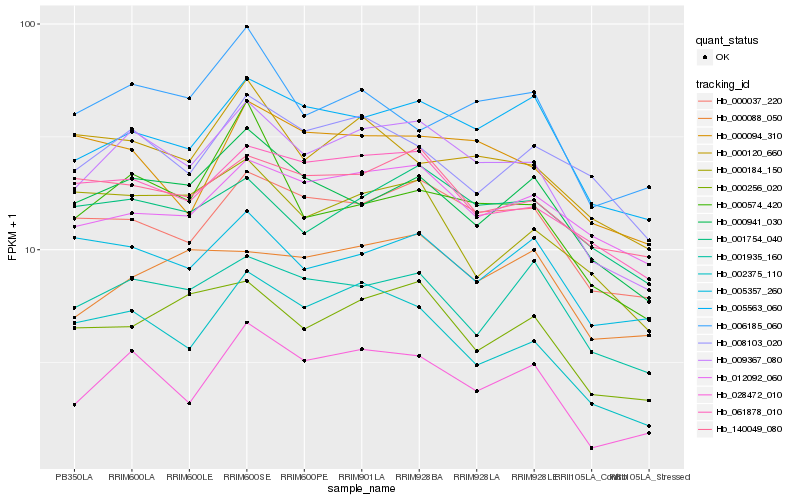
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009367_080 |
0.0 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 42 [Jatropha curcas] |
| 2 |
Hb_028472_010 |
0.0844073013 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_000037_220 |
0.0876350584 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 4 |
Hb_061878_010 |
0.0926261438 |
- |
- |
hypothetical protein CISIN_1g0094072mg, partial [Citrus sinensis] |
| 5 |
Hb_005563_060 |
0.0933955857 |
- |
- |
PREDICTED: coatomer subunit alpha-1 [Jatropha curcas] |
| 6 |
Hb_002375_110 |
0.0948834264 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 7 |
Hb_000256_020 |
0.098420319 |
transcription factor |
TF Family: PHD |
PREDICTED: CHD3-type chromatin-remodeling factor PICKLE isoform X1 [Jatropha curcas] |
| 8 |
Hb_000941_030 |
0.0991074122 |
- |
- |
PREDICTED: uncharacterized protein LOC105646531 [Jatropha curcas] |
| 9 |
Hb_001935_160 |
0.1070243281 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 10 |
Hb_140049_080 |
0.1071573598 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 11 |
Hb_012092_060 |
0.1110461326 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 12 |
Hb_001754_040 |
0.1115741856 |
transcription factor |
TF Family: TRAF |
PREDICTED: uncharacterized protein LOC105643434 [Jatropha curcas] |
| 13 |
Hb_000574_420 |
0.1117316805 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |
| 14 |
Hb_000120_660 |
0.1121339771 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 15 |
Hb_008103_020 |
0.1141817755 |
- |
- |
hypothetical protein POPTR_0003s20310g [Populus trichocarpa] |
| 16 |
Hb_005357_260 |
0.1149754458 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 17 |
Hb_000088_050 |
0.1153261242 |
- |
- |
PREDICTED: threonylcarbamoyladenosine tRNA methylthiotransferase [Jatropha curcas] |
| 18 |
Hb_000184_150 |
0.115952811 |
- |
- |
PREDICTED: U-box domain-containing protein 15 [Jatropha curcas] |
| 19 |
Hb_006185_060 |
0.1159957473 |
- |
- |
PREDICTED: protein phosphatase 2C 16 [Jatropha curcas] |
| 20 |
Hb_000094_310 |
0.1169732399 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g75140 [Jatropha curcas] |