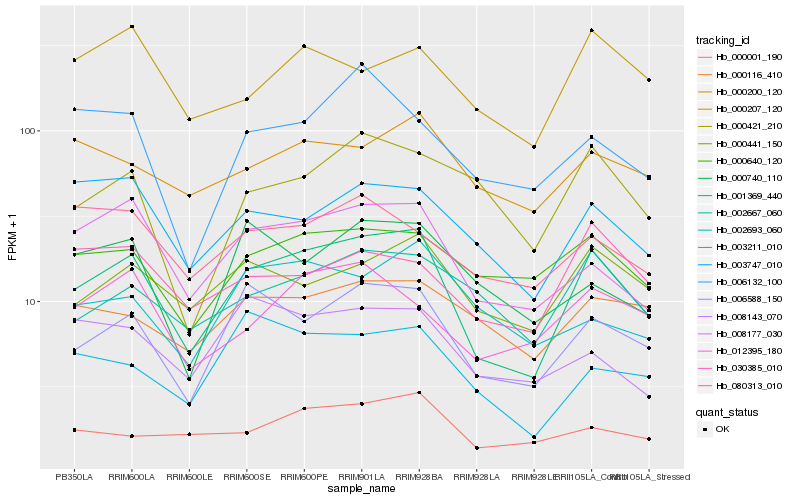
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_440 |
0.0 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 7 [Jatropha curcas] |
| 2 |
Hb_006588_150 |
0.1194137969 |
- |
- |
PREDICTED: steroid 5-alpha-reductase DET2 [Jatropha curcas] |
| 3 |
Hb_008177_030 |
0.1314249237 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_000441_150 |
0.1474788222 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, root-type isozyme, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_002667_060 |
0.1511887132 |
- |
- |
mannose-1-phosphate guanyltransferase, putative [Ricinus communis] |
| 6 |
Hb_003747_010 |
0.1525025577 |
- |
- |
hypothetical protein JCGZ_24080 [Jatropha curcas] |
| 7 |
Hb_000640_120 |
0.1543115066 |
- |
- |
PREDICTED: uncharacterized protein LOC105642625 [Jatropha curcas] |
| 8 |
Hb_012395_180 |
0.1579965645 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |
| 9 |
Hb_003211_010 |
0.1608058508 |
- |
- |
PREDICTED: uncharacterized protein LOC105635705 [Jatropha curcas] |
| 10 |
Hb_000421_210 |
0.1616078919 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000001_190 |
0.1616397017 |
- |
- |
hypothetical protein CISIN_1g0082522mg, partial [Citrus sinensis] |
| 12 |
Hb_000200_120 |
0.1651143368 |
- |
- |
PREDICTED: probable fructose-bisphosphate aldolase 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000207_120 |
0.1656365326 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.3 [Jatropha curcas] |
| 14 |
Hb_000116_410 |
0.1667686135 |
- |
- |
hypothetical protein JCGZ_22416 [Jatropha curcas] |
| 15 |
Hb_006132_100 |
0.1674072383 |
- |
- |
PREDICTED: uncharacterized protein LOC105642968 [Jatropha curcas] |
| 16 |
Hb_008143_070 |
0.1674375088 |
- |
- |
PREDICTED: factor of DNA methylation 4 [Jatropha curcas] |
| 17 |
Hb_002693_060 |
0.1693093509 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_030385_010 |
0.1694773065 |
- |
- |
PREDICTED: zinc-binding alcohol dehydrogenase domain-containing protein 2-like [Jatropha curcas] |
| 19 |
Hb_080313_010 |
0.1694818304 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 20 |
Hb_000740_110 |
0.170323384 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01850 [Jatropha curcas] |