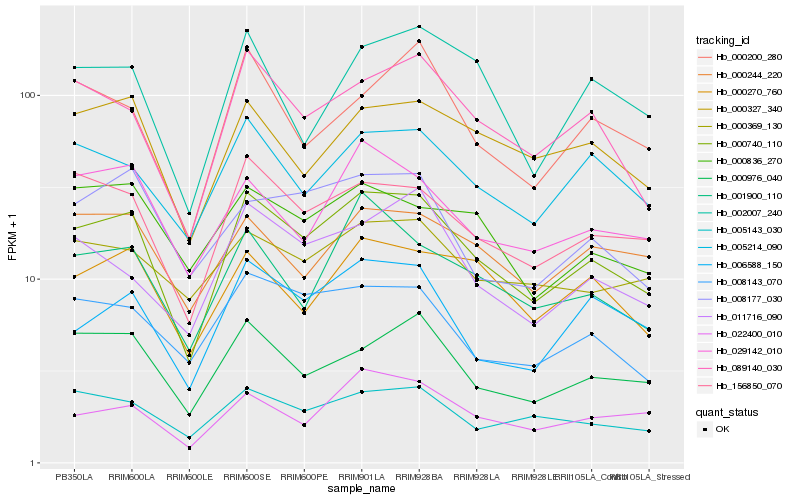
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000740_110 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01850 [Jatropha curcas] |
| 2 |
Hb_156850_070 |
0.0922429176 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_089140_030 |
0.0986346589 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |
| 4 |
Hb_000976_040 |
0.1080686095 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 5 |
Hb_008143_070 |
0.1089234416 |
- |
- |
PREDICTED: factor of DNA methylation 4 [Jatropha curcas] |
| 6 |
Hb_006588_150 |
0.1101419802 |
- |
- |
PREDICTED: steroid 5-alpha-reductase DET2 [Jatropha curcas] |
| 7 |
Hb_005214_090 |
0.1112668735 |
- |
- |
PREDICTED: uncharacterized protein LOC105636015 [Jatropha curcas] |
| 8 |
Hb_000327_340 |
0.1121725144 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 9 |
Hb_005143_030 |
0.1142897442 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 10 |
Hb_000270_760 |
0.1162466139 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_029142_010 |
0.1167711672 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 12 |
Hb_001900_110 |
0.1168240747 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit E [Jatropha curcas] |
| 13 |
Hb_022400_010 |
0.1179007791 |
- |
- |
Nitrilase/cyanide hydratase and apolipoprotein N-acyltransferase family protein [Theobroma cacao] |
| 14 |
Hb_011716_090 |
0.1181609223 |
transcription factor |
TF Family: HRT |
putative transciption factor family protein [Populus trichocarpa] |
| 15 |
Hb_000244_220 |
0.1223574786 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_000836_270 |
0.1232252691 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2 [Jatropha curcas] |
| 17 |
Hb_008177_030 |
0.1233251649 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_000200_280 |
0.1255287741 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 19 |
Hb_000369_130 |
0.1255462831 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 20 |
Hb_002007_240 |
0.1257139737 |
- |
- |
PREDICTED: F-box protein At1g55000 [Jatropha curcas] |