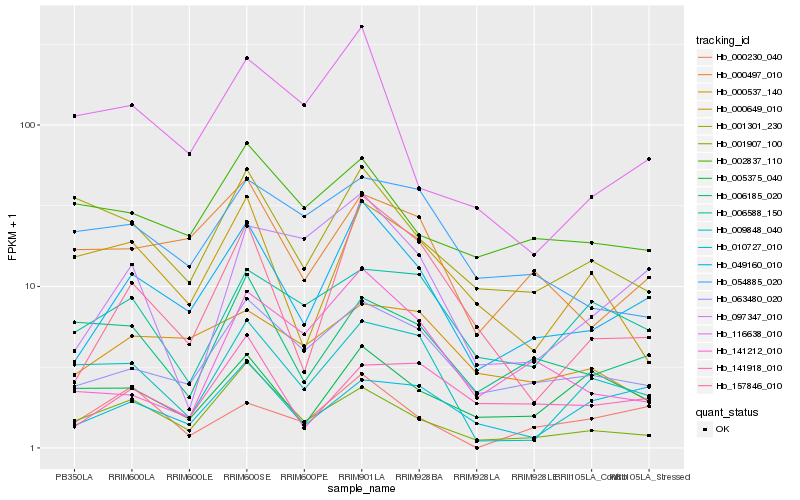
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_049160_010 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 2 |
Hb_063480_020 |
0.1516232346 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_157846_010 |
0.1595920871 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 4 |
Hb_000497_010 |
0.1810546643 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHR2 [Jatropha curcas] |
| 5 |
Hb_141918_010 |
0.1955377231 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 6 |
Hb_001907_100 |
0.2001982879 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 7 |
Hb_010727_010 |
0.2022188289 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 8 |
Hb_097347_010 |
0.2035715639 |
- |
- |
PREDICTED: molybdenum cofactor sulfurase-like, partial [Malus domestica] |
| 9 |
Hb_000537_140 |
0.212141233 |
- |
- |
lupus la ribonucleoprotein, putative [Ricinus communis] |
| 10 |
Hb_006185_020 |
0.2129713284 |
- |
- |
PREDICTED: midasin [Jatropha curcas] |
| 11 |
Hb_002837_110 |
0.2160459639 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 2, mitochondrial [Jatropha curcas] |
| 12 |
Hb_005375_040 |
0.2164427474 |
- |
- |
dicer-1, putative [Ricinus communis] |
| 13 |
Hb_141212_010 |
0.2182296788 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 14 |
Hb_116638_010 |
0.2187636168 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000649_010 |
0.2209153154 |
- |
- |
PREDICTED: adagio protein 3 [Jatropha curcas] |
| 16 |
Hb_001301_230 |
0.2219132827 |
- |
- |
PREDICTED: uncharacterized protein LOC105632949 isoform X2 [Jatropha curcas] |
| 17 |
Hb_006588_150 |
0.2230284086 |
- |
- |
PREDICTED: steroid 5-alpha-reductase DET2 [Jatropha curcas] |
| 18 |
Hb_009848_040 |
0.2230434924 |
- |
- |
hypothetical protein B456_009G349600 [Gossypium raimondii] |
| 19 |
Hb_054885_020 |
0.2260380818 |
- |
- |
- |
| 20 |
Hb_000230_040 |
0.2266345304 |
- |
- |
PREDICTED: protein MON2 homolog isoform X2 [Vitis vinifera] |