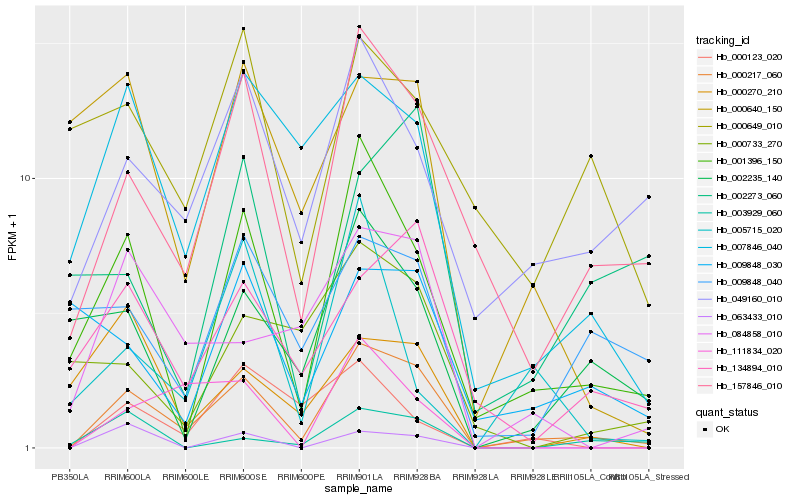
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000217_060 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_157846_010 |
0.2175913972 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 3 |
Hb_001396_150 |
0.220843519 |
transcription factor |
TF Family: M-type |
hypothetical protein POPTR_0001s13660g [Populus trichocarpa] |
| 4 |
Hb_049160_010 |
0.2642960701 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 5 |
Hb_084858_010 |
0.2681871458 |
- |
- |
Uncharacterized protein isoform 3 [Theobroma cacao] |
| 6 |
Hb_007846_040 |
0.2695076678 |
- |
- |
PREDICTED: uncharacterized protein LOC105646691 [Jatropha curcas] |
| 7 |
Hb_134894_010 |
0.2719828247 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like isoform X2 [Gossypium raimondii] |
| 8 |
Hb_111834_020 |
0.293474616 |
- |
- |
PREDICTED: protein phosphatase methylesterase 1 [Jatropha curcas] |
| 9 |
Hb_002235_140 |
0.2963844729 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003929_060 |
0.2982067962 |
- |
- |
PREDICTED: uncharacterized protein ECU03_1610 [Jatropha curcas] |
| 11 |
Hb_000123_020 |
0.3005825173 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At1g61190 [Vitis vinifera] |
| 12 |
Hb_063433_010 |
0.3069366418 |
- |
- |
Chloroplast channel forming outer membrane protein isoform 2 [Theobroma cacao] |
| 13 |
Hb_000640_150 |
0.3152841346 |
transcription factor |
TF Family: WRKY |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_009848_040 |
0.3155665149 |
- |
- |
hypothetical protein B456_009G349600 [Gossypium raimondii] |
| 15 |
Hb_000733_270 |
0.3223963851 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1-alpha catalytic subunit [Dasypus novemcinctus] |
| 16 |
Hb_000270_210 |
0.3236096723 |
- |
- |
PREDICTED: transcription initiation factor IIB-2-like [Jatropha curcas] |
| 17 |
Hb_000649_010 |
0.3241556284 |
- |
- |
PREDICTED: adagio protein 3 [Jatropha curcas] |
| 18 |
Hb_005715_020 |
0.3245967319 |
- |
- |
PREDICTED: ras-related protein RABC2a-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_002273_060 |
0.3247924952 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_009848_030 |
0.3251212608 |
- |
- |
long-chain-fatty-acid CoA ligase, putative [Ricinus communis] |