| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
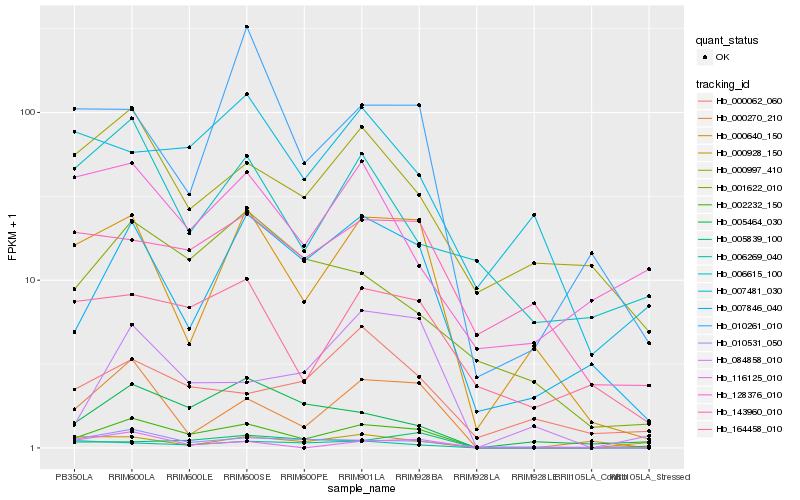
Hb_002232_150 |
0.0 |
- |
- |
PREDICTED: squalene monooxygenase-like [Jatropha curcas] |
| 2 |
Hb_007846_040 |
0.2036271975 |
- |
- |
PREDICTED: uncharacterized protein LOC105646691 [Jatropha curcas] |
| 3 |
Hb_000270_210 |
0.2046264078 |
- |
- |
PREDICTED: transcription initiation factor IIB-2-like [Jatropha curcas] |
| 4 |
Hb_010531_050 |
0.2080369788 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000640_150 |
0.2147377732 |
transcription factor |
TF Family: WRKY |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005464_030 |
0.2195360323 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_164458_010 |
0.2381273181 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_006269_040 |
0.2392389171 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000928_150 |
0.2393176553 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001622_010 |
0.2414957348 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_084858_010 |
0.2454000823 |
- |
- |
Uncharacterized protein isoform 3 [Theobroma cacao] |
| 12 |
Hb_005839_100 |
0.2458587098 |
- |
- |
PREDICTED: APO protein 1, chloroplastic isoform X3 [Jatropha curcas] |
| 13 |
Hb_000997_410 |
0.2467603984 |
- |
- |
PREDICTED: uncharacterized protein LOC105632066 [Jatropha curcas] |
| 14 |
Hb_116125_010 |
0.2569659013 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710-like [Citrus sinensis] |
| 15 |
Hb_143960_010 |
0.2584099433 |
- |
- |
- |
| 16 |
Hb_128376_010 |
0.2599592644 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_006615_100 |
0.2604539225 |
- |
- |
PREDICTED: uncharacterized protein LOC105640683 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000062_060 |
0.262171523 |
transcription factor |
TF Family: Orphans |
multi-sensor hybrid histidine kinase [Plautia stali symbiont] |
| 19 |
Hb_007481_030 |
0.2640593805 |
- |
- |
PREDICTED: uncharacterized protein LOC105628433 [Jatropha curcas] |
| 20 |
Hb_010261_010 |
0.2653072937 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |