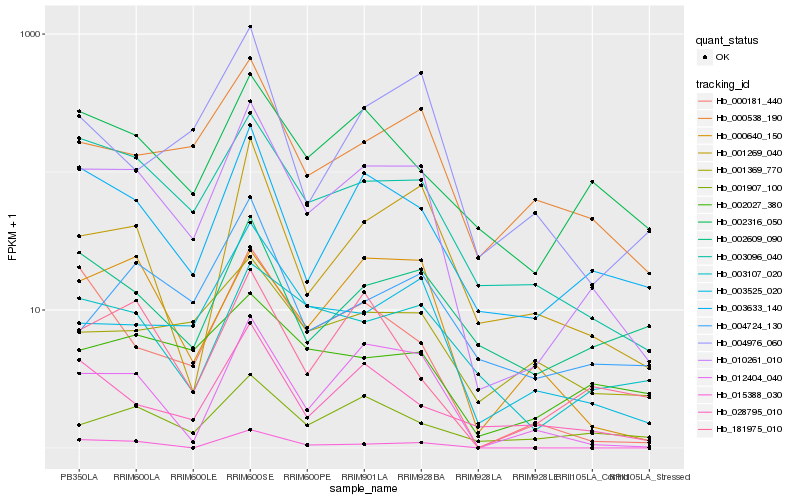
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010261_010 |
0.0 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 2 |
Hb_003096_040 |
0.1546122792 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0005s07290g [Populus trichocarpa] |
| 3 |
Hb_003633_140 |
0.1625996877 |
- |
- |
PREDICTED: uncharacterized protein LOC105647486 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003525_020 |
0.1639445948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_012404_040 |
0.1722891572 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 2-2, chloroplastic/amyloplastic [Jatropha curcas] |
| 6 |
Hb_000538_190 |
0.1795886383 |
- |
- |
PREDICTED: protein DJ-1 homolog D [Jatropha curcas] |
| 7 |
Hb_001269_040 |
0.1893475577 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_002316_050 |
0.1948750392 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001907_100 |
0.1993309561 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 10 |
Hb_003107_020 |
0.2002169084 |
- |
- |
hypothetical protein JCGZ_18656 [Jatropha curcas] |
| 11 |
Hb_000640_150 |
0.2023656442 |
transcription factor |
TF Family: WRKY |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_015388_030 |
0.2025795415 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_028795_010 |
0.2047821202 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 14 |
Hb_000181_440 |
0.2056565175 |
- |
- |
- |
| 15 |
Hb_002609_090 |
0.2083212168 |
- |
- |
PREDICTED: phosphomethylpyrimidine synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_004724_130 |
0.2088522813 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14 [Jatropha curcas] |
| 17 |
Hb_001369_770 |
0.2096674206 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 18 |
Hb_181975_010 |
0.2111734067 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Populus euphratica] |
| 19 |
Hb_002027_380 |
0.212082446 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase isoform X2 [Jatropha curcas] |
| 20 |
Hb_004976_060 |
0.2125534571 |
- |
- |
- |