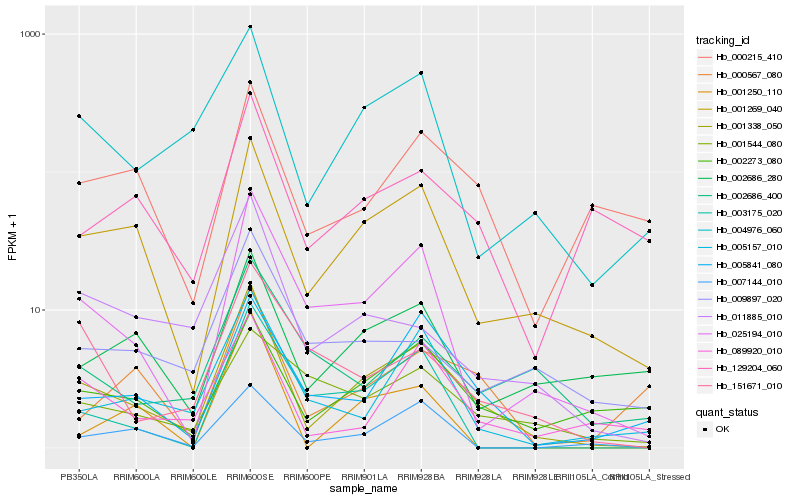
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001338_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0016s05400g [Populus trichocarpa] |
| 2 |
Hb_001269_040 |
0.1788123297 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_025194_010 |
0.1879064279 |
- |
- |
PREDICTED: uncharacterized protein LOC105644857 [Jatropha curcas] |
| 4 |
Hb_004976_060 |
0.2108281238 |
- |
- |
- |
| 5 |
Hb_005157_010 |
0.2183140487 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 6 |
Hb_005841_080 |
0.2187840493 |
- |
- |
hypothetical protein POPTR_0018s11370g [Populus trichocarpa] |
| 7 |
Hb_089920_010 |
0.2196468483 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 8 |
Hb_011885_010 |
0.22224432 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 9 |
Hb_002273_080 |
0.2244495884 |
- |
- |
geranylgeranyl reductase [Monoraphidium neglectum] |
| 10 |
Hb_003175_020 |
0.2307869733 |
- |
- |
homeobox protein knotted-1, putative [Ricinus communis] |
| 11 |
Hb_007144_010 |
0.238930391 |
- |
- |
- |
| 12 |
Hb_151671_010 |
0.2468454478 |
- |
- |
hypothetical protein POPTR_0005s04441g [Populus trichocarpa] |
| 13 |
Hb_009897_020 |
0.24699434 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 14 |
Hb_002686_280 |
0.2516054793 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_001250_110 |
0.2527182676 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0010s05340g [Populus trichocarpa] |
| 16 |
Hb_000567_080 |
0.2534615077 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |
| 17 |
Hb_000215_410 |
0.2537856193 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 18 |
Hb_002686_400 |
0.2545161994 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 19 |
Hb_129204_060 |
0.2545826943 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 20 |
Hb_001544_080 |
0.2570948694 |
- |
- |
PREDICTED: 66 kDa stress protein-like [Jatropha curcas] |