| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
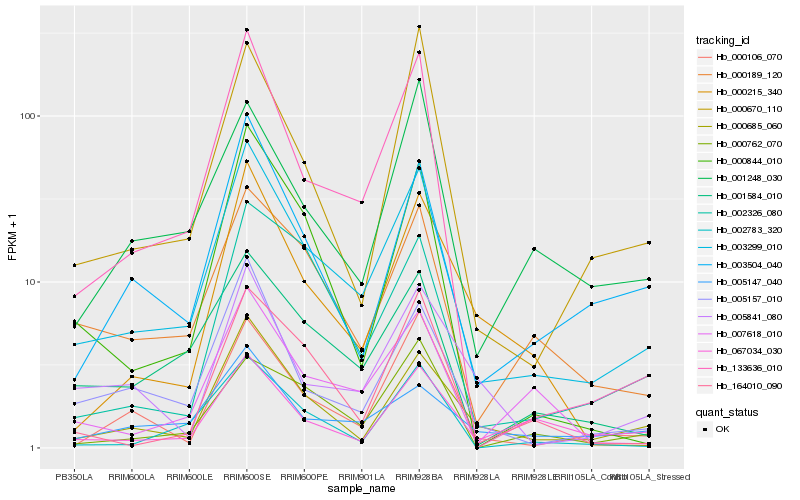
Hb_003299_010 |
0.0 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 2 |
Hb_005157_010 |
0.1399769996 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 3 |
Hb_000685_060 |
0.1403839406 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 4 |
Hb_000189_120 |
0.1433060565 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like isoform X5 [Jatropha curcas] |
| 5 |
Hb_001584_010 |
0.1434803189 |
- |
- |
kelch repeat protein, putative [Ricinus communis] |
| 6 |
Hb_003504_040 |
0.1501982595 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 2-like [Jatropha curcas] |
| 7 |
Hb_133636_010 |
0.1518380677 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Brassica rapa] |
| 8 |
Hb_007618_010 |
0.1618511857 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 9 |
Hb_002326_080 |
0.1705236961 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.6 [Jatropha curcas] |
| 10 |
Hb_005147_040 |
0.1715515833 |
- |
- |
PREDICTED: uncharacterized protein LOC105629674 [Jatropha curcas] |
| 11 |
Hb_002783_320 |
0.1721476595 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FC isoform X1 [Jatropha curcas] |
| 12 |
Hb_000106_070 |
0.1725825451 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 13 |
Hb_000670_110 |
0.1741140705 |
- |
- |
l-asparaginase, putative [Ricinus communis] |
| 14 |
Hb_000844_010 |
0.1783599832 |
- |
- |
PREDICTED: lysine histidine transporter-like 8 [Jatropha curcas] |
| 15 |
Hb_000762_070 |
0.1799642042 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 16 |
Hb_001248_030 |
0.1804918067 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_164010_090 |
0.1827775612 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 18 |
Hb_067034_030 |
0.1860096729 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 19 |
Hb_000215_340 |
0.1895117974 |
- |
- |
gamma glutamyl transpeptidases, putative [Ricinus communis] |
| 20 |
Hb_005841_080 |
0.1895172596 |
- |
- |
hypothetical protein POPTR_0018s11370g [Populus trichocarpa] |