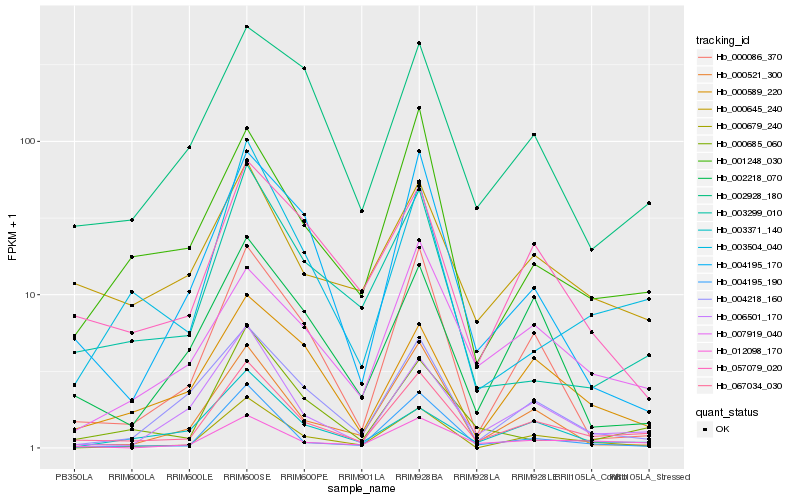
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_067034_030 |
0.0 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 2 |
Hb_012098_170 |
0.1431321471 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_16874 [Jatropha curcas] |
| 3 |
Hb_000679_240 |
0.1561282117 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 4 |
Hb_006501_170 |
0.1562909752 |
- |
- |
hypothetical protein JCGZ_07371 [Jatropha curcas] |
| 5 |
Hb_001248_030 |
0.1609438424 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_000645_240 |
0.165588467 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 7 |
Hb_000521_300 |
0.1686861615 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 8 |
Hb_007919_040 |
0.1690381691 |
- |
- |
PREDICTED: protein LYK5 [Jatropha curcas] |
| 9 |
Hb_003504_040 |
0.1721254574 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 2-like [Jatropha curcas] |
| 10 |
Hb_004195_190 |
0.172382301 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 11 |
Hb_000086_370 |
0.1751659179 |
- |
- |
Long-chain-alcohol oxidase FAO2 [Triticum urartu] |
| 12 |
Hb_002928_180 |
0.1774786421 |
- |
- |
catalase CAT1 [Manihot esculenta] |
| 13 |
Hb_004218_160 |
0.1789054338 |
- |
- |
NBS-LRR type disease resistance protein [Medicago truncatula] |
| 14 |
Hb_057079_020 |
0.1797018332 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000589_220 |
0.1814980706 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 16 |
Hb_000685_060 |
0.1834846818 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 17 |
Hb_003299_010 |
0.1860096729 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 18 |
Hb_004195_170 |
0.1889721767 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_003371_140 |
0.1891829685 |
- |
- |
hypothetical protein POPTR_0001s23540g, partial [Populus trichocarpa] |
| 20 |
Hb_002218_070 |
0.1902610489 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |