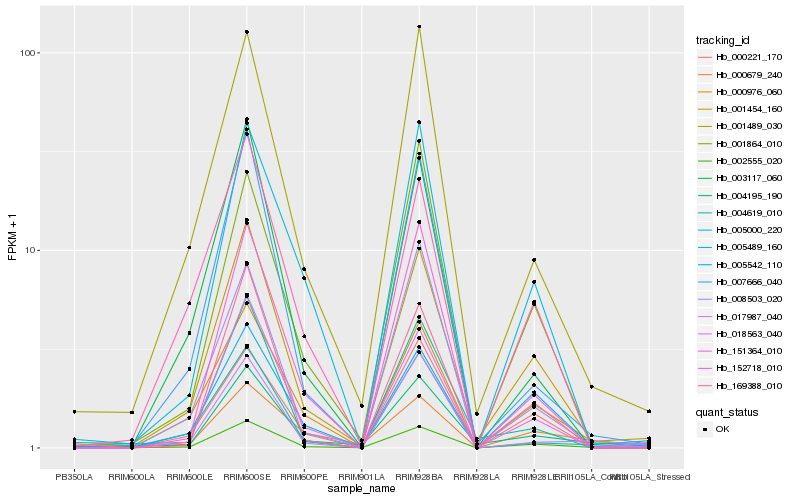
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002555_020 |
0.0 |
- |
- |
Receptor-like protein kinase [Medicago truncatula] |
| 2 |
Hb_001454_160 |
0.0788915379 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 3 |
Hb_018563_040 |
0.1122592762 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_004195_190 |
0.1191726496 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 5 |
Hb_000976_060 |
0.1258574296 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 6 |
Hb_001489_030 |
0.1274159901 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.6 [Jatropha curcas] |
| 7 |
Hb_005542_110 |
0.1398762013 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01203 [Jatropha curcas] |
| 8 |
Hb_007666_040 |
0.1410606771 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_017987_040 |
0.144514073 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0002s11440g [Populus trichocarpa] |
| 10 |
Hb_005489_160 |
0.1484361796 |
- |
- |
PREDICTED: primary amine oxidase-like [Jatropha curcas] |
| 11 |
Hb_169388_010 |
0.1504427523 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 12 |
Hb_151364_010 |
0.150492295 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 13 |
Hb_005000_220 |
0.1528188534 |
- |
- |
gulonolactone oxidase, putative [Ricinus communis] |
| 14 |
Hb_001864_010 |
0.1562551732 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000221_170 |
0.1570487213 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 2 [Jatropha curcas] |
| 16 |
Hb_008503_020 |
0.1578165228 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 17 |
Hb_003117_060 |
0.1596514691 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 18 |
Hb_152718_010 |
0.1610564944 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 19 |
Hb_000679_240 |
0.1630886232 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 20 |
Hb_004619_010 |
0.1632646962 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |