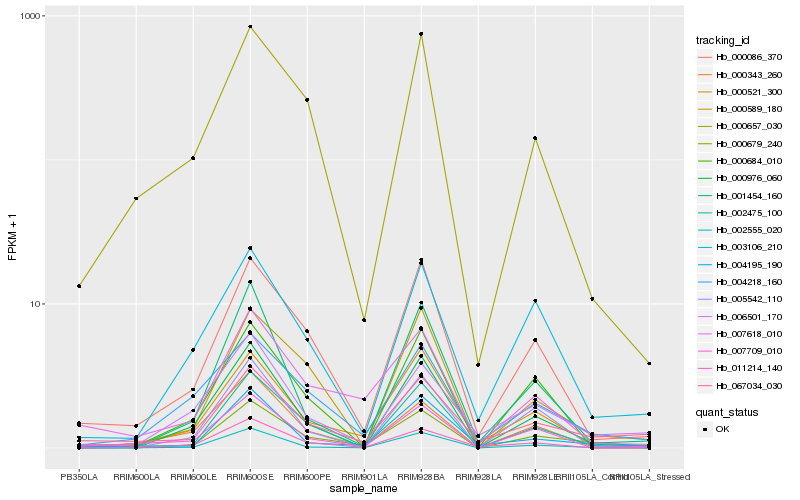
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000679_240 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 2 |
Hb_067034_030 |
0.1561282117 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 3 |
Hb_000589_180 |
0.1601353902 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 4 |
Hb_000521_300 |
0.1611696537 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 5 |
Hb_002555_020 |
0.1630886232 |
- |
- |
Receptor-like protein kinase [Medicago truncatula] |
| 6 |
Hb_004195_190 |
0.1648223585 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 7 |
Hb_000086_370 |
0.166744337 |
- |
- |
Long-chain-alcohol oxidase FAO2 [Triticum urartu] |
| 8 |
Hb_000976_060 |
0.1677822037 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 9 |
Hb_005542_110 |
0.1757565159 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01203 [Jatropha curcas] |
| 10 |
Hb_002475_100 |
0.178373081 |
- |
- |
PREDICTED: uncharacterized protein LOC105645451 [Jatropha curcas] |
| 11 |
Hb_011214_140 |
0.1788933232 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 12 |
Hb_004218_160 |
0.1789360633 |
- |
- |
NBS-LRR type disease resistance protein [Medicago truncatula] |
| 13 |
Hb_000657_030 |
0.1844082452 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 14 |
Hb_006501_170 |
0.1870969424 |
- |
- |
hypothetical protein JCGZ_07371 [Jatropha curcas] |
| 15 |
Hb_003106_210 |
0.18946121 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74B1-like [Jatropha curcas] |
| 16 |
Hb_000343_260 |
0.189478371 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 17 |
Hb_007618_010 |
0.1910793666 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 18 |
Hb_007709_010 |
0.1919803667 |
- |
- |
- |
| 19 |
Hb_001454_160 |
0.1921653941 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000684_010 |
0.1994021729 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |