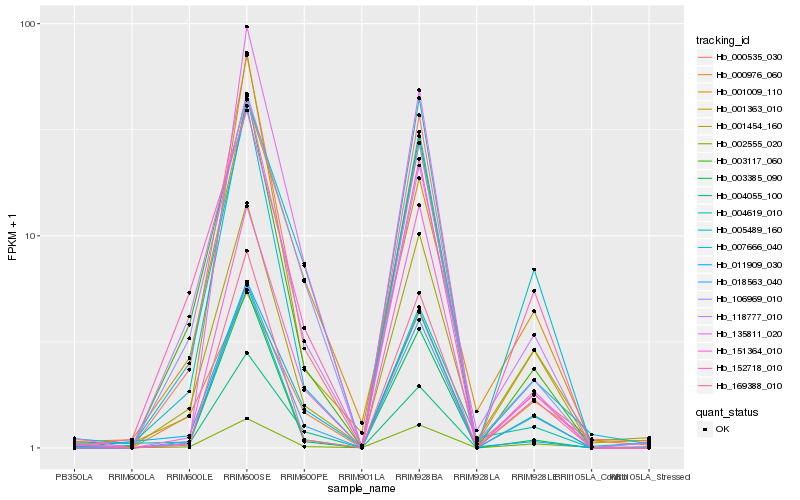
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018563_040 |
0.0 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_001454_160 |
0.1014079339 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002555_020 |
0.1122592762 |
- |
- |
Receptor-like protein kinase [Medicago truncatula] |
| 4 |
Hb_169388_010 |
0.1139566124 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 5 |
Hb_151364_010 |
0.1267280924 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 6 |
Hb_003385_090 |
0.1286473616 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 7 |
Hb_003117_060 |
0.1301645234 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 8 |
Hb_007666_040 |
0.1307373874 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_118777_010 |
0.130756298 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000535_030 |
0.1327863762 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 11 |
Hb_005489_160 |
0.1355838985 |
- |
- |
PREDICTED: primary amine oxidase-like [Jatropha curcas] |
| 12 |
Hb_106969_010 |
0.1401715188 |
- |
- |
Cytochrome P450 76C4 [Glycine soja] |
| 13 |
Hb_000976_060 |
0.1469290686 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 14 |
Hb_011909_030 |
0.1484379972 |
- |
- |
hypothetical protein JCGZ_00732 [Jatropha curcas] |
| 15 |
Hb_001363_010 |
0.1500821368 |
- |
- |
hypothetical protein CISIN_1g027763mg [Citrus sinensis] |
| 16 |
Hb_004055_100 |
0.1516269277 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 17 |
Hb_135811_020 |
0.1528225879 |
- |
- |
hypothetical protein JCGZ_19965 [Jatropha curcas] |
| 18 |
Hb_152718_010 |
0.1528480492 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 19 |
Hb_004619_010 |
0.1538625708 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 20 |
Hb_001009_110 |
0.1540228397 |
- |
- |
PREDICTED: uncharacterized protein LOC105640232 [Jatropha curcas] |