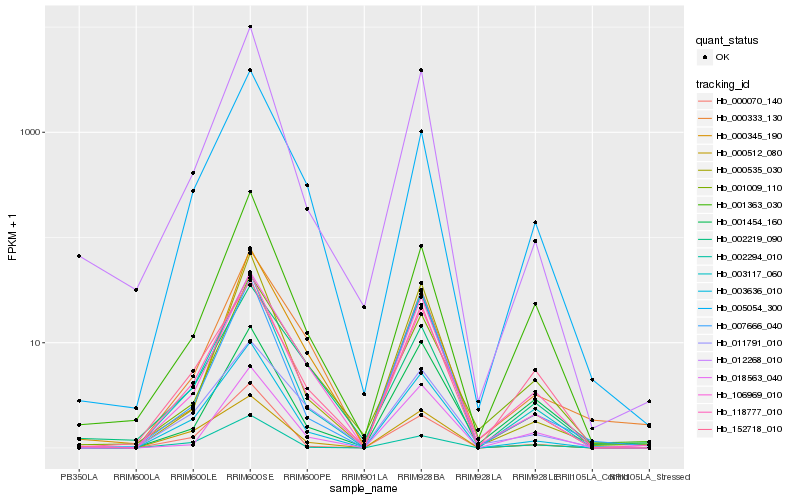
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_118777_010 |
0.0 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_003636_010 |
0.0854669428 |
- |
- |
hypothetical protein CISIN_1g037850mg, partial [Citrus sinensis] |
| 3 |
Hb_003117_060 |
0.0967717234 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 4 |
Hb_152718_010 |
0.0989165919 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 5 |
Hb_001363_030 |
0.1065973539 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F2-like [Populus euphratica] |
| 6 |
Hb_106969_010 |
0.1067331139 |
- |
- |
Cytochrome P450 76C4 [Glycine soja] |
| 7 |
Hb_000070_140 |
0.1147593931 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 8 |
Hb_001009_110 |
0.1176692001 |
- |
- |
PREDICTED: uncharacterized protein LOC105640232 [Jatropha curcas] |
| 9 |
Hb_005054_300 |
0.1207385904 |
- |
- |
aquaporin [Hevea brasiliensis] |
| 10 |
Hb_011791_010 |
0.1277659268 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 11 |
Hb_012268_010 |
0.1296721256 |
- |
- |
glutamic acid-rich protein [Manihot esculenta] |
| 12 |
Hb_000333_130 |
0.1302805182 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_018563_040 |
0.130756298 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_000535_030 |
0.1326098187 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 15 |
Hb_002294_010 |
0.1329441858 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 16 |
Hb_000512_080 |
0.1337957182 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_001454_160 |
0.135806733 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 18 |
Hb_002219_090 |
0.1371019866 |
- |
- |
protein kinase atmrk1, putative [Ricinus communis] |
| 19 |
Hb_000345_190 |
0.1410857149 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0005s02730g [Populus trichocarpa] |
| 20 |
Hb_007666_040 |
0.1423645629 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic-like [Jatropha curcas] |