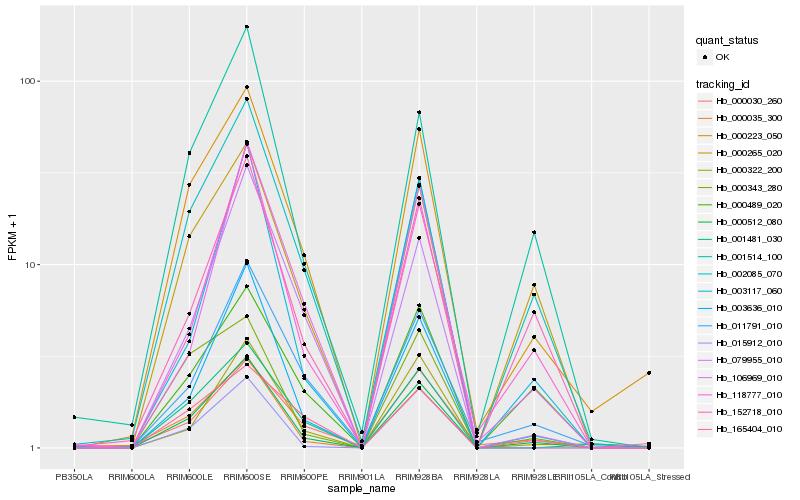
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000512_080 |
0.0 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_003636_010 |
0.0974876919 |
- |
- |
hypothetical protein CISIN_1g037850mg, partial [Citrus sinensis] |
| 3 |
Hb_000223_050 |
0.1058900929 |
- |
- |
PREDICTED: endochitinase A-like [Jatropha curcas] |
| 4 |
Hb_152718_010 |
0.1272758343 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 5 |
Hb_001514_100 |
0.128496677 |
- |
- |
PREDICTED: F-box protein GID2 [Jatropha curcas] |
| 6 |
Hb_000265_020 |
0.1307233188 |
- |
- |
hypothetical protein POPTR_0011s16980g, partial [Populus trichocarpa] |
| 7 |
Hb_002085_070 |
0.1316068683 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_000035_300 |
0.1326206807 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_118777_010 |
0.1337957182 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_003117_060 |
0.1346163453 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 11 |
Hb_011791_010 |
0.1360750173 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 12 |
Hb_015912_010 |
0.1373118658 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 13 |
Hb_001481_030 |
0.139643519 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_000343_280 |
0.141528693 |
- |
- |
hypothetical protein JCGZ_24094 [Jatropha curcas] |
| 15 |
Hb_000322_200 |
0.1440448281 |
transcription factor |
TF Family: bHLH |
PREDICTED: putative transcription factor bHLH041 [Jatropha curcas] |
| 16 |
Hb_079955_010 |
0.1492733015 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 17 |
Hb_000030_260 |
0.1499699288 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 18 |
Hb_000489_020 |
0.1500837126 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Populus euphratica] |
| 19 |
Hb_106969_010 |
0.1554345743 |
- |
- |
Cytochrome P450 76C4 [Glycine soja] |
| 20 |
Hb_165404_010 |
0.1555876621 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |