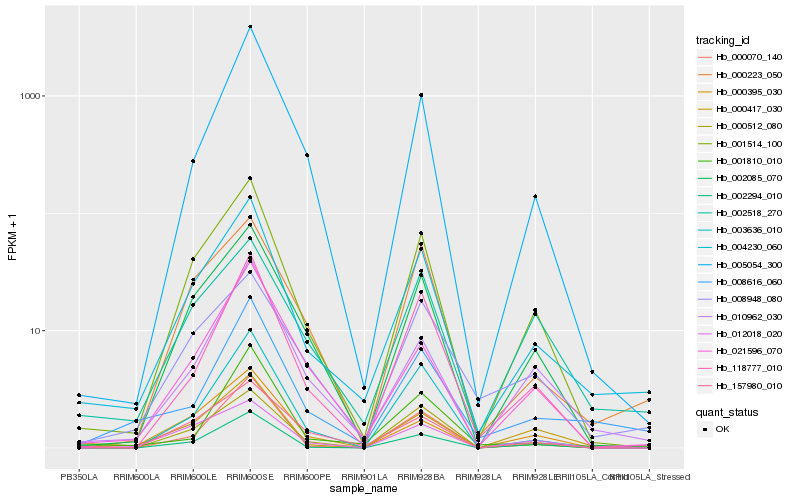
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004230_060 |
0.0 |
- |
- |
PREDICTED: ferritin, chloroplastic-like [Populus euphratica] |
| 2 |
Hb_001514_100 |
0.1157289353 |
- |
- |
PREDICTED: F-box protein GID2 [Jatropha curcas] |
| 3 |
Hb_002085_070 |
0.1443836575 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 4 |
Hb_000417_030 |
0.1460672465 |
transcription factor |
TF Family: G2-like |
PREDICTED: putative two-component response regulator ARR19 [Jatropha curcas] |
| 5 |
Hb_008616_060 |
0.1468746582 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 6 |
Hb_000223_050 |
0.1513622722 |
- |
- |
PREDICTED: endochitinase A-like [Jatropha curcas] |
| 7 |
Hb_010962_030 |
0.1546750788 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_000395_030 |
0.1555850007 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 9 |
Hb_002294_010 |
0.1571428321 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 10 |
Hb_002518_270 |
0.1593995349 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 11 |
Hb_118777_010 |
0.1650797702 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_008948_080 |
0.1654727765 |
- |
- |
PREDICTED: TBC1 domain family member 13-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000512_080 |
0.1671645131 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_021596_070 |
0.1672112832 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 15 |
Hb_157980_010 |
0.1683174436 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 16 |
Hb_012018_020 |
0.1700303229 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Eucalyptus grandis] |
| 17 |
Hb_005054_300 |
0.1708757257 |
- |
- |
aquaporin [Hevea brasiliensis] |
| 18 |
Hb_003636_010 |
0.1711653677 |
- |
- |
hypothetical protein CISIN_1g037850mg, partial [Citrus sinensis] |
| 19 |
Hb_000070_140 |
0.1742806154 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 20 |
Hb_001810_010 |
0.1752272511 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Nelumbo nucifera] |