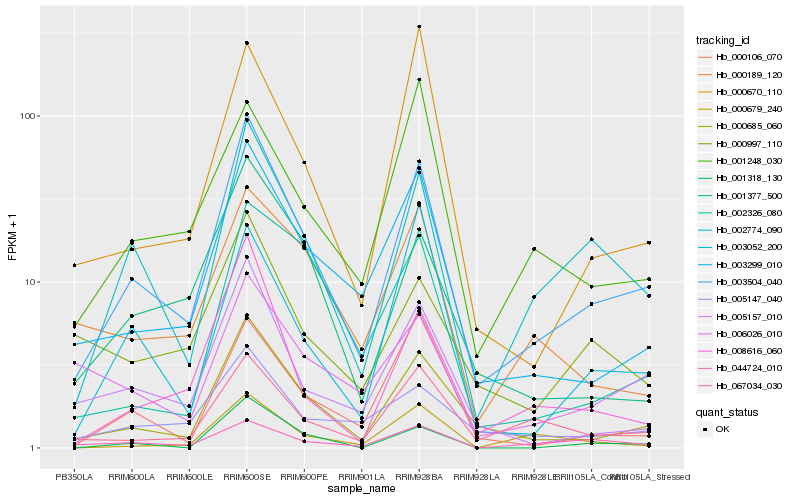
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003504_040 |
0.0 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 2-like [Jatropha curcas] |
| 2 |
Hb_000685_060 |
0.1171595893 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 3 |
Hb_002774_090 |
0.1381021993 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003299_010 |
0.1501982595 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 5 |
Hb_005157_010 |
0.1705345434 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 6 |
Hb_067034_030 |
0.1721254574 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 7 |
Hb_008616_060 |
0.1734423852 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 8 |
Hb_001377_500 |
0.1834478228 |
- |
- |
DOF AFFECTING GERMINATION 2 family protein [Populus trichocarpa] |
| 9 |
Hb_005147_040 |
0.1841709224 |
- |
- |
PREDICTED: uncharacterized protein LOC105629674 [Jatropha curcas] |
| 10 |
Hb_044724_010 |
0.1915262906 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 11 |
Hb_000106_070 |
0.1984137982 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 12 |
Hb_001318_130 |
0.2004846073 |
- |
- |
PREDICTED: short-chain dehydrogenase reductase 3b-like [Populus euphratica] |
| 13 |
Hb_000670_110 |
0.2026137136 |
- |
- |
l-asparaginase, putative [Ricinus communis] |
| 14 |
Hb_001248_030 |
0.2034205984 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_000189_120 |
0.2049105912 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like isoform X5 [Jatropha curcas] |
| 16 |
Hb_000997_110 |
0.205461908 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 17 |
Hb_003052_200 |
0.2079583032 |
- |
- |
hypothetical protein POPTR_0010s14360g [Populus trichocarpa] |
| 18 |
Hb_006026_010 |
0.2089850687 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 3-like [Jatropha curcas] |
| 19 |
Hb_002326_080 |
0.2124361625 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.6 [Jatropha curcas] |
| 20 |
Hb_000679_240 |
0.2133824993 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |