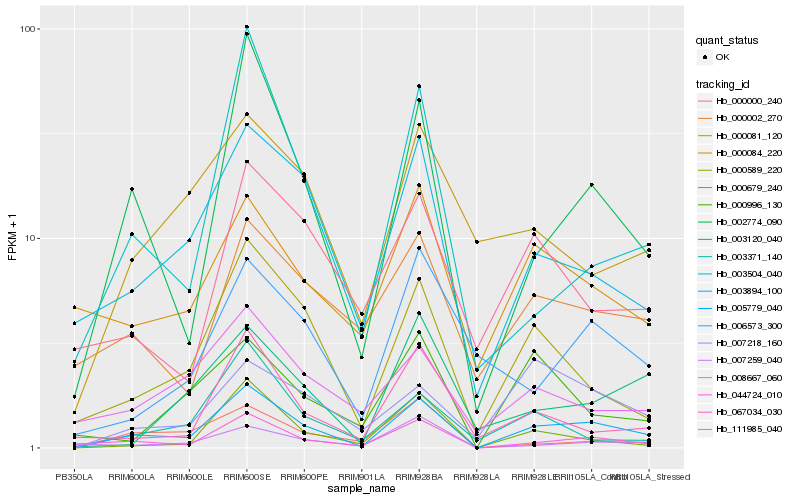
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005779_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_044724_010 |
0.1712754629 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 3 |
Hb_002774_090 |
0.1742733818 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000589_220 |
0.2020147633 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 5 |
Hb_003894_100 |
0.2098724209 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1-like [Jatropha curcas] |
| 6 |
Hb_000679_240 |
0.22803183 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 7 |
Hb_067034_030 |
0.2319720107 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 8 |
Hb_003504_040 |
0.2406671943 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 2-like [Jatropha curcas] |
| 9 |
Hb_007218_160 |
0.2406804757 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 10 |
Hb_008667_060 |
0.2429578379 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 11 |
Hb_000002_270 |
0.2433412424 |
- |
- |
hypothetical protein VITISV_043897 [Vitis vinifera] |
| 12 |
Hb_003371_140 |
0.2439229341 |
- |
- |
hypothetical protein POPTR_0001s23540g, partial [Populus trichocarpa] |
| 13 |
Hb_003120_040 |
0.2447162081 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 [Jatropha curcas] |
| 14 |
Hb_007259_040 |
0.2461887788 |
- |
- |
hypothetical protein VITISV_005492 [Vitis vinifera] |
| 15 |
Hb_000081_120 |
0.2467286077 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 16 |
Hb_000000_240 |
0.2486293528 |
- |
- |
PREDICTED: protein LYK5-like [Jatropha curcas] |
| 17 |
Hb_006573_300 |
0.248667566 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_111985_040 |
0.2507751577 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 3 [Jatropha curcas] |
| 19 |
Hb_000084_220 |
0.2530627542 |
- |
- |
PREDICTED: uncharacterized protein LOC105634692 [Jatropha curcas] |
| 20 |
Hb_000996_130 |
0.2537335195 |
- |
- |
PREDICTED: remorin-like [Populus euphratica] |