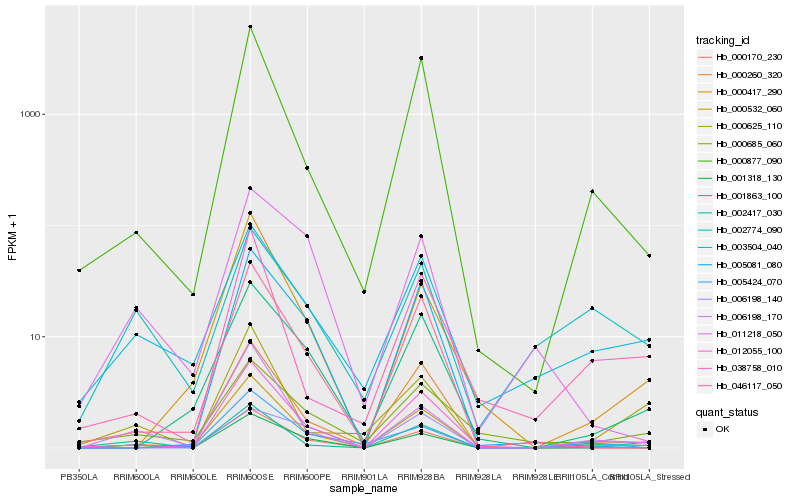
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001318_130 |
0.0 |
- |
- |
PREDICTED: short-chain dehydrogenase reductase 3b-like [Populus euphratica] |
| 2 |
Hb_006198_170 |
0.1916572125 |
- |
- |
PREDICTED: serine/threonine-protein kinase At3g07070-like [Jatropha curcas] |
| 3 |
Hb_012055_100 |
0.1928124315 |
- |
- |
heat shock protein 70 (HSP70)-interacting protein, putative [Ricinus communis] |
| 4 |
Hb_003504_040 |
0.2004846073 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 2-like [Jatropha curcas] |
| 5 |
Hb_001863_100 |
0.2201169964 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002774_090 |
0.2203082234 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_006198_140 |
0.2211807998 |
- |
- |
hypothetical protein L484_015208 [Morus notabilis] |
| 8 |
Hb_000260_320 |
0.2233939313 |
- |
- |
PREDICTED: uncharacterized protein LOC105649058 [Jatropha curcas] |
| 9 |
Hb_000877_090 |
0.2253865574 |
- |
- |
allergenic-related protein Pt2L4 [Manihot esculenta] |
| 10 |
Hb_000170_230 |
0.226924967 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000685_060 |
0.2299881425 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 12 |
Hb_011218_050 |
0.2315569755 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 13 |
Hb_000417_290 |
0.2353985168 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005424_070 |
0.2358063988 |
- |
- |
PREDICTED: TATA-box-binding protein 1-like [Jatropha curcas] |
| 15 |
Hb_002417_030 |
0.2360869781 |
- |
- |
PREDICTED: purine permease 3-like [Jatropha curcas] |
| 16 |
Hb_000625_110 |
0.2393396842 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 17 |
Hb_046117_050 |
0.2438630012 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0009s14590g [Populus trichocarpa] |
| 18 |
Hb_005081_080 |
0.2450726493 |
- |
- |
hypothetical protein JCGZ_16787 [Jatropha curcas] |
| 19 |
Hb_000532_060 |
0.2455654938 |
- |
- |
copper-transporting atpase p-type, putative [Ricinus communis] |
| 20 |
Hb_038758_010 |
0.2462988931 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |