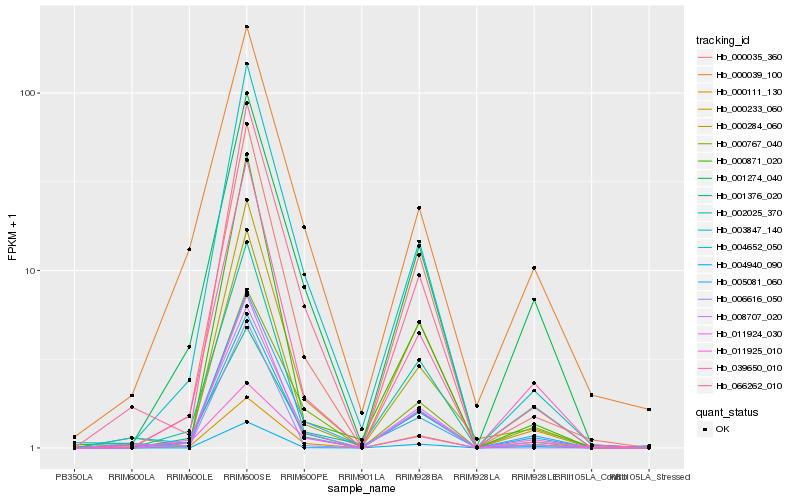
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004652_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s15360g [Populus trichocarpa] |
| 2 |
Hb_066262_010 |
0.1195032459 |
- |
- |
hypothetical protein JCGZ_14798 [Jatropha curcas] |
| 3 |
Hb_000233_060 |
0.1203584048 |
- |
- |
hypothetical protein JCGZ_14793 [Jatropha curcas] |
| 4 |
Hb_039650_010 |
0.1226284087 |
- |
- |
cellulose synthase-like protein [Populus tomentosa] |
| 5 |
Hb_008707_020 |
0.1255242904 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_000284_060 |
0.130188612 |
desease resistance |
Gene Name: NB-ARC |
disease resistance protein (TIR-NBS-LRR class), putative [Medicago truncatula] |
| 7 |
Hb_000767_040 |
0.133065055 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 8 |
Hb_002025_370 |
0.1340188026 |
- |
- |
monooxygenase, putative [Ricinus communis] |
| 9 |
Hb_001274_040 |
0.1382608578 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |
| 10 |
Hb_011924_030 |
0.1390851219 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_000035_360 |
0.139877989 |
- |
- |
PREDICTED: VQ motif-containing protein 31-like [Jatropha curcas] |
| 12 |
Hb_001376_020 |
0.1412745956 |
- |
- |
hypothetical protein JCGZ_01547 [Jatropha curcas] |
| 13 |
Hb_003847_140 |
0.1427830102 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 14 |
Hb_005081_060 |
0.1507820996 |
- |
- |
hypothetical protein EUGRSUZ_C00833 [Eucalyptus grandis] |
| 15 |
Hb_006616_050 |
0.1518453672 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 16 |
Hb_000111_130 |
0.1521847794 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 17 |
Hb_004940_090 |
0.153444197 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 18 |
Hb_011925_010 |
0.1561204282 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000039_100 |
0.1563024303 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 20 |
Hb_000871_020 |
0.1579798189 |
- |
- |
receptor-kinase, putative [Ricinus communis] |