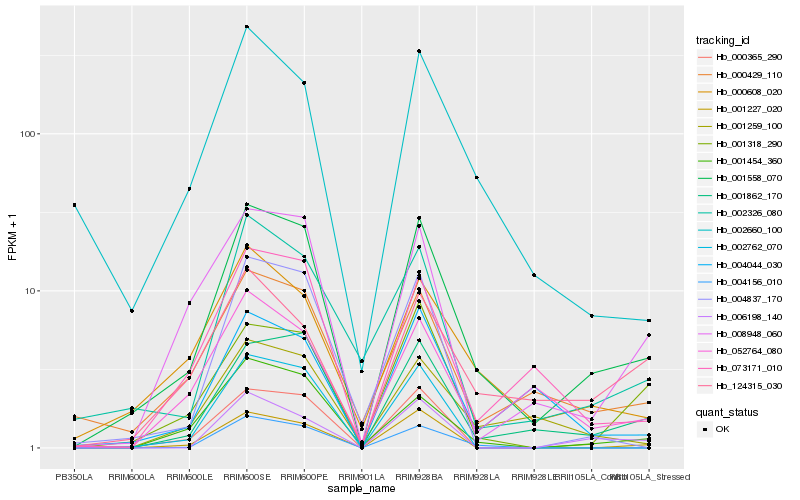
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001558_070 |
0.0 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 2 |
Hb_001862_170 |
0.1404904344 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_000608_020 |
0.1617834003 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 4 |
Hb_001454_360 |
0.1655728481 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET17 [Jatropha curcas] |
| 5 |
Hb_000429_110 |
0.1683912896 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein WIP2-like [Populus euphratica] |
| 6 |
Hb_008948_060 |
0.1702186723 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_073171_010 |
0.1739095938 |
- |
- |
PREDICTED: ABC transporter G family member 29-like [Jatropha curcas] |
| 8 |
Hb_001318_290 |
0.1758884976 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 9 |
Hb_001259_100 |
0.1772066759 |
- |
- |
chloride channel-like family protein [Populus trichocarpa] |
| 10 |
Hb_002326_080 |
0.177800693 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.6 [Jatropha curcas] |
| 11 |
Hb_001227_020 |
0.1811625844 |
- |
- |
- |
| 12 |
Hb_002660_100 |
0.1888982083 |
- |
- |
plasma membrane intrinsic protein [Hevea brasiliensis] |
| 13 |
Hb_004156_010 |
0.1898135334 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 14 |
Hb_124315_030 |
0.1903399307 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 15 |
Hb_004044_030 |
0.19321528 |
- |
- |
PREDICTED: uncharacterized protein LOC105639335 [Jatropha curcas] |
| 16 |
Hb_052764_080 |
0.1939566877 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD5 isoform X1 [Jatropha curcas] |
| 17 |
Hb_006198_140 |
0.1970049666 |
- |
- |
hypothetical protein L484_015208 [Morus notabilis] |
| 18 |
Hb_004837_170 |
0.1982260982 |
- |
- |
PREDICTED: uncharacterized protein LOC105648284 [Jatropha curcas] |
| 19 |
Hb_002762_070 |
0.1998029603 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 20 |
Hb_000365_290 |
0.2029057532 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |