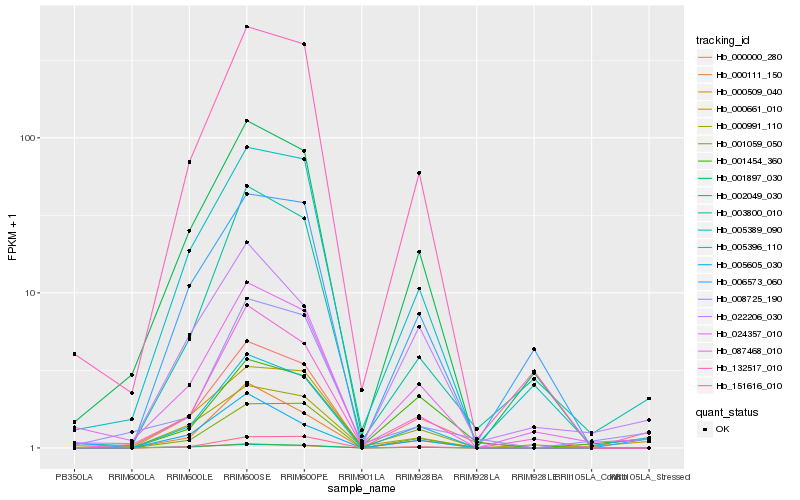
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000000_280 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005396_110 |
0.1525971978 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000509_040 |
0.1564859348 |
- |
- |
hypothetical protein VITISV_009716 [Vitis vinifera] |
| 4 |
Hb_003800_010 |
0.164014664 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |
| 5 |
Hb_132517_010 |
0.1694399523 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like isoform X3 [Populus euphratica] |
| 6 |
Hb_001454_360 |
0.171065665 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET17 [Jatropha curcas] |
| 7 |
Hb_000661_010 |
0.1757828315 |
- |
- |
peroxisomal 3-keto-acyl-CoA thiolase [Hevea brasiliensis] |
| 8 |
Hb_005389_090 |
0.1761757703 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA27, putative [Ricinus communis] |
| 9 |
Hb_001897_030 |
0.1795826087 |
- |
- |
PREDICTED: actin-depolymerizing factor 5 [Jatropha curcas] |
| 10 |
Hb_001059_050 |
0.1796608299 |
- |
- |
PREDICTED: putative F-box/LRR-repeat protein At3g18150 [Jatropha curcas] |
| 11 |
Hb_024357_010 |
0.1825084182 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 12 |
Hb_000111_150 |
0.1829816603 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 13 |
Hb_008725_190 |
0.1855173301 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 5-like [Jatropha curcas] |
| 14 |
Hb_002049_030 |
0.1889086048 |
- |
- |
PREDICTED: uncharacterized protein LOC105635940 isoform X3 [Jatropha curcas] |
| 15 |
Hb_022206_030 |
0.1926183877 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 16 |
Hb_000991_110 |
0.1941777939 |
- |
- |
PREDICTED: uncharacterized protein LOC105636336 [Jatropha curcas] |
| 17 |
Hb_151616_010 |
0.1953365426 |
- |
- |
- |
| 18 |
Hb_087468_010 |
0.1960030672 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 19 |
Hb_005605_030 |
0.1961556352 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 20 |
Hb_006573_060 |
0.1977669939 |
- |
- |
hypothetical protein [Bruguiera gymnorhiza] |