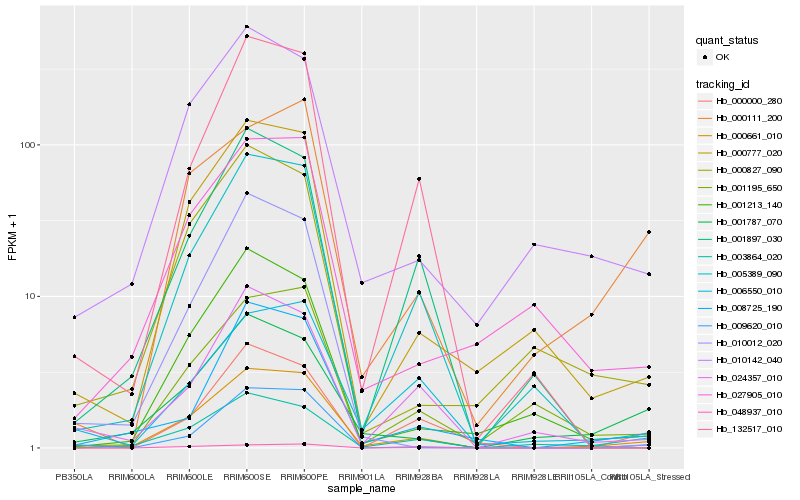
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000661_010 |
0.0 |
- |
- |
peroxisomal 3-keto-acyl-CoA thiolase [Hevea brasiliensis] |
| 2 |
Hb_000777_020 |
0.1349067762 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11 [Jatropha curcas] |
| 3 |
Hb_005389_090 |
0.1354508354 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA27, putative [Ricinus communis] |
| 4 |
Hb_009620_010 |
0.1499785518 |
transcription factor |
TF Family: Orphans |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_010142_040 |
0.1543704876 |
- |
- |
PREDICTED: classical arabinogalactan protein 4-like [Jatropha curcas] |
| 6 |
Hb_132517_010 |
0.1547748246 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like isoform X3 [Populus euphratica] |
| 7 |
Hb_008725_190 |
0.1605547466 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 5-like [Jatropha curcas] |
| 8 |
Hb_024357_010 |
0.1618144179 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 9 |
Hb_006550_010 |
0.1664080587 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Eucalyptus grandis] |
| 10 |
Hb_001195_650 |
0.1664493827 |
- |
- |
PREDICTED: calcium uniporter protein 4, mitochondrial [Jatropha curcas] |
| 11 |
Hb_001213_140 |
0.16799506 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 12 |
Hb_000827_090 |
0.1689483856 |
- |
- |
PREDICTED: uncharacterized protein LOC103714121 [Phoenix dactylifera] |
| 13 |
Hb_001787_070 |
0.1708345548 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Camelina sativa] |
| 14 |
Hb_001897_030 |
0.1710869569 |
- |
- |
PREDICTED: actin-depolymerizing factor 5 [Jatropha curcas] |
| 15 |
Hb_027905_010 |
0.1713150238 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 16 |
Hb_000111_200 |
0.1747033373 |
- |
- |
PREDICTED: CYSTM1 family protein A-like [Cucumis melo] |
| 17 |
Hb_048937_010 |
0.1748771524 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IX.1 [Nelumbo nucifera] |
| 18 |
Hb_000000_280 |
0.1757828315 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003864_020 |
0.1767287232 |
- |
- |
kinase, putative [Ricinus communis] |
| 20 |
Hb_010012_020 |
0.1778885311 |
- |
- |
conserved hypothetical protein [Ricinus communis] |